5N01
 
 | | Crystal structure of the decarboxylase AibA/AibB C56N variant | | Descriptor: | ACETATE ION, GLYCEROL, Glutaconate CoA-transferase family, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NCJ
 
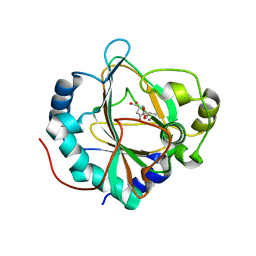 | | GriE in complex with manganese, succinate and (2S,4R)-5-hydroxyleucine | | Descriptor: | (2S,4R)-5-hydroxyleucine, Leucine hydroxylase, MANGANESE (II) ION, ... | | Authors: | Lukat, P, Blankenfeldt, W, Mueller, R. | | Deposit date: | 2017-03-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity.
Chem Sci, 8, 2017
|
|
5MZX
 
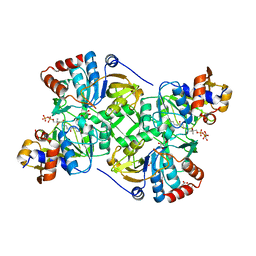 | | Crystal structure of the decarboxylase AibA/AibB in complex with 4'-diphospho pantetheine | | Descriptor: | 4'-diphospho pantetheine, ACETATE ION, GLYCEROL, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6ZSU
 
 | |
6ZSV
 
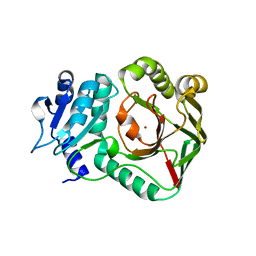 | | Structure of crocagin biosynthetic protein CgnB | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Koehnke, J, Adam, S. | | Deposit date: | 2020-07-16 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unusual peptide-binding proteins guide pyrroloindoline alkaloid formation in crocagin biosynthesis.
Nat.Chem., 15, 2023
|
|
5HQW
 
 | | Crystal structure of a trans-AT PKS dehydratase domain of C0ZGQ6 from Brevibacillus brevis | | Descriptor: | MAGNESIUM ION, Putative polyketide synthase | | Authors: | Jakob, R.P, Hauswirth, P, Dilmi, J, Herbst, D.A, Maier, T. | | Deposit date: | 2016-01-22 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structures of Dehydratase Domains from trans AT Polyketide Biosynthetic Pathway.
To Be Published
|
|
5HWP
 
 | | MvaS with acetylated Cys115 in complex with coenzyme A | | Descriptor: | COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
5HWO
 
 | | MvaS in complex with 3-hydroxy-3-methylglutaryl coenzyme A | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
5HST
 
 | |
5HWQ
 
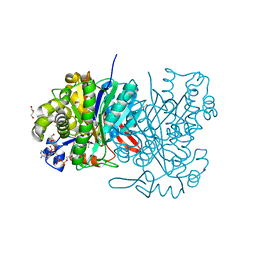 | | MvaS in complex with acetoacetyl coenzyme A | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
5HWR
 
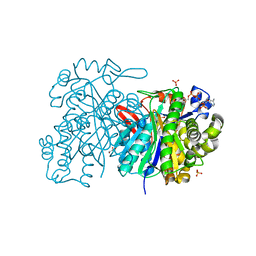 | | MvaS in complex with coenzyme A | | Descriptor: | COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
5IL6
 
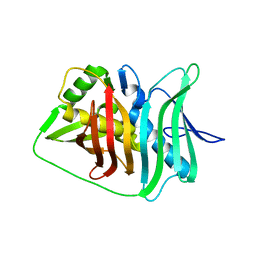 | |
5IL5
 
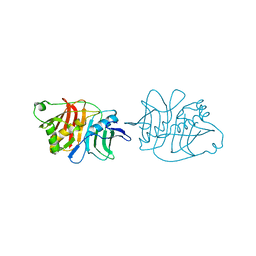 | |
3E1T
 
 | | Structure and action of the myxobacterial chondrochloren halogenase CndH, a new variant of FAD-dependent halogenases | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Halogenase | | Authors: | Buedenbender, S, Rachid, S, Mueller, R, Schulz, G.E. | | Deposit date: | 2008-08-04 | | Release date: | 2009-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and action of the myxobacterial chondrochloren halogenase CndH: a new variant of FAD-dependent halogenases.
J.Mol.Biol., 385, 2009
|
|
5J6O
 
 | |
5HU7
 
 | |
7OSE
 
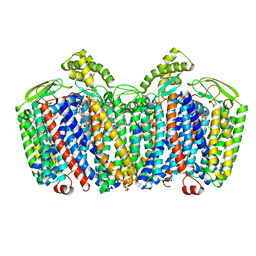 | | cytochrome bd-II type oxidase with bound aurachin D | | Descriptor: | Aurachin D, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-II ubiquinol oxidase subunit 1, ... | | Authors: | Grauel, A, Kaegi, J, Rasmussen, T, Wohlwend, D, Boettcher, B, Friedrich, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Escherichia coli cytochrome bd-II type oxidase with bound aurachin D.
Nat Commun, 12, 2021
|
|
7OTC
 
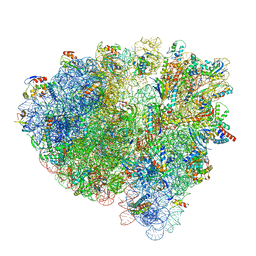 | | Cryo-EM structure of an Escherichia coli 70S ribosome in complex with elongation factor G and the antibiotic Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Wieland, M, Koller, T.O, Wilson, D.N. | | Deposit date: | 2021-06-10 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4GSA
 
 | |
7PD7
 
 | | Crocagin methyl transferase CgnL | | Descriptor: | GLYCEROL, Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zheng, D, Koehnke, J. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Unusual peptide-binding proteins guide pyrroloindoline alkaloid formation in crocagin biosynthesis.
Nat.Chem., 15, 2023
|
|
5LHK
 
 | | Bottromycin maturation enzyme BotP in complex with Mn | | Descriptor: | BICARBONATE ION, Leucine aminopeptidase 2, chloroplastic, ... | | Authors: | Koehnke, J, Adam, S. | | Deposit date: | 2016-07-12 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Substrate Recognition of the Bottromycin Maturation Enzyme BotP.
Chembiochem, 17, 2016
|
|
8A2N
 
 | | Structure of crocagin biosynthetic protein CgnD | | Descriptor: | CgnD, SULFATE ION | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Unusual peptide-binding proteins guide pyrroloindoline alkaloid formation in crocagin biosynthesis.
Nat.Chem., 15, 2023
|
|
8ADG
 
 | | Cryo-EM structure of Darobactin 22 bound BAM complex | | Descriptor: | Darobactin 22, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Yuan, B, Marlovits, T.C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ADI
 
 | | Cryo-EM structure of Darobactin 9 bound BAM complex | | Descriptor: | Darobactin 9, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Yuan, B, Marlovits, T.C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5LHJ
 
 | | Bottromycin maturation enzyme BotP | | Descriptor: | CHLORIDE ION, GLYCEROL, Leucine aminopeptidase 2, ... | | Authors: | Koehnke, J, Mann, G. | | Deposit date: | 2016-07-12 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and Substrate Recognition of the Bottromycin Maturation Enzyme BotP.
Chembiochem, 17, 2016
|
|
