4KXW
 
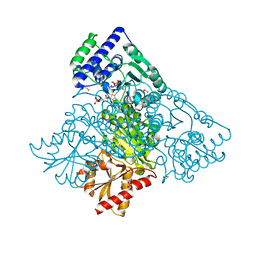 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 2 | | Descriptor: | 1,2-ETHANEDIOL, D-XYLITOL-5-PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4KXV
 
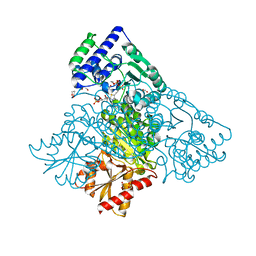 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D-XYLITOL-5-PHOSPHATE, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4KGD
 
 | |
5HSA
 
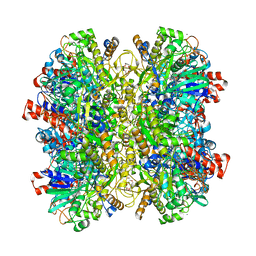 | | Alcohol Oxidase AOX1 from Pichia Pastoris | | Descriptor: | ARABINO-FLAVIN-ADENINE DINUCLEOTIDE, Alcohol oxidase 1, CALCIUM ION, ... | | Authors: | Neumann, P, Ficner, R, Feussner, I, Koch, C. | | Deposit date: | 2016-01-25 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Alcohol Oxidase from Pichia pastoris.
Plos One, 11, 2016
|
|
3CGN
 
 | | Crystal Structure of thermophilic SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, SULFATE ION | | Authors: | Neumann, P, Loew, C, Stubbs, M.T, Balbach, J. | | Deposit date: | 2008-03-06 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure Determination and Functional Characterization of the Metallochaperone SlyD from Thermus thermophilus
J.Mol.Biol., 398, 2010
|
|
3FT7
 
 | | Crystal structure of an extremely stable dimeric protein from sulfolobus islandicus | | Descriptor: | GLYCEROL, Uncharacterized protein ORF56 | | Authors: | Neumann, P, Loew, C, Weininger, U, Stubbs, M.T. | | Deposit date: | 2009-01-12 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Stability Analysis of an Extremely Stable Dimeric DNA Binding Protein from Sulfolobus islandicus
Biochemistry, 48, 2009
|
|
5L4X
 
 | | Crystal structure of FimH lectin domain in complex with 4-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 4-deoxy-4-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4U
 
 | | Crystal structure of FimH lectin domain in complex with 2-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 2-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4Y
 
 | | Crystal structure of FimH lectin domain in complex with 4-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 4-deoxy-4-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4V
 
 | | Crystal structure of FimH lectin domain in complex with 3-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 3-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4T
 
 | | Crystal structure of FimH lectin domain in complex with 2-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 2-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
8ARU
 
 | |
3JZI
 
 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3UWI
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-02 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UPE
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | 1-[(7-carbamimidoylnaphthalen-2-yl)methyl]-6-({1-[(1Z)-ethanimidoyl]piperidin-4-yl}oxy)-2-(propan-2-yl)-1H-indole-4-carboxylic acid, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-18 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UQV
 
 | | Bovine trypsin variant X(triplePhe227) in complex with small molecule inhibitor | | Descriptor: | 3-(3-carbamimidoylphenyl)-N-(2'-sulfamoylbiphenyl-4-yl)-1,2-oxazole-4-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3UY9
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4FEE
 
 | | High-resolution structure of pyruvate oxidase in complex with reaction intermediate 2-hydroxyethyl-thiamin diphosphate carbanion-enamine, crystal B | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Meyer, D, Neumann, P, Koers, E, Sjuts, H, Luedtke, S, Sheldrick, G.M, Ficner, R, Tittmann, K. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Unexpected tautomeric equilibria of the carbanion-enamine intermediate in pyruvate oxidase highlight unrecognized chemical versatility of thiamin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FEG
 
 | | High-resolution structure of pyruvate oxidase in complex with reaction intermediate 2-hydroxyethyl-thiamin diphosphate carbanion-enamine, crystal A | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Meyer, D, Neumann, P, Koers, E, Sjuts, H, Luedtke, S, Sheldrick, G.M, Ficner, R, Tittmann, K. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Unexpected tautomeric equilibria of the carbanion-enamine intermediate in pyruvate oxidase highlight unrecognized chemical versatility of thiamin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4O1J
 
 | | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora. | | Descriptor: | CHLORIDE ION, Carbonic anhydrase, ZINC ION | | Authors: | Lehneck, R, Neumann, P, Vullo, D, Elleuche, S, Supuran, C.T, Ficner, R, Poggeler, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora.
Febs J., 281, 2014
|
|
2FYI
 
 | | Crystal Structure of the Cofactor-Binding Domain of the Cbl Transcriptional Regulator | | Descriptor: | HTH-type transcriptional regulator cbl | | Authors: | Stec, E, Neumann, P, Wilkinson, A.J, Brzozowski, A.M, Bujacz, G.D. | | Deposit date: | 2006-02-08 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of the Sulphate Starvation Response in E. coli: Crystal Structure and Mutational Analysis of the Cofactor-binding Domain of the Cbl Transcriptional Regulator.
J.Mol.Biol., 364, 2006
|
|
6Z3X
 
 | | Crystal structure of the designed antibody DesAb-anti-HSA-P1 | | Descriptor: | CACODYLATE ION, DesAb-anti-HSA-P1, IMIDAZOLE, ... | | Authors: | Costanzi, E, Sormanni, P, Ricagno, S. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based computational design of antibodies targeting structured epitopes.
Sci Adv, 8, 2022
|
|
6ZXA
 
 | | LH2 complex from Marichromatium purpuratum | | Descriptor: | 9-cis-okenone, BACTERIOCHLOROPHYLL A, LHC domain-containing protein, ... | | Authors: | Gardiner, A.T, Naydenova, K, Castro-Hartmann, P, Nguyen-Phan, T.C, Russo, C.J, Sader, K, Hunter, C.N, Cogdell, R.J, Qian, P. | | Deposit date: | 2020-07-29 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | The 2.4 angstrom cryo-EM structure of a heptameric light-harvesting 2 complex reveals two carotenoid energy transfer pathways.
Sci Adv, 7, 2021
|
|
4RLE
 
 | | Crystal structure of the c-di-AMP binding PII-like protein DarA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, NICKEL (II) ION, Uncharacterized protein YaaQ | | Authors: | Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2014-10-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification, Characterization, and Structure Analysis of the Cyclic di-AMP-binding PII-like Signal Transduction Protein DarA.
J.Biol.Chem., 290, 2015
|
|
4XRI
 
 | | Crystal Structure of Importin Beta in an Ammonium Sulfate Condition | | Descriptor: | GLYCEROL, Putative uncharacterized protein, SULFATE ION | | Authors: | Tauchert, M.J, Neumann, P, Ficner, R, Dickmanns, A. | | Deposit date: | 2015-01-21 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Impact of the crystallization condition on importin-beta conformation.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
