1FV5
 
 | | SOLUTION STRUCTURE OF THE FIRST ZINC FINGER FROM THE DROSOPHILA U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | FIRST ZINC FINGER OF U-SHAPED, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-18 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
1FU9
 
 | | SOLUTION STRUCTURE OF THE NINTH ZINC-FINGER DOMAIN OF THE U-SHAPED TRANSCRIPTION FACTOR | | Descriptor: | U-SHAPED TRANSCRIPTIONAL COFACTOR, ZINC ION | | Authors: | Liew, C.K, Kowalski, K, Fox, A.H, Newton, A, Sharpe, B.K, Crossley, M, Mackay, J.P. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein-protein interactions.
Structure Fold.Des., 8, 2000
|
|
1K2F
 
 | | siah, Seven In Absentia Homolog | | Descriptor: | BETA-MERCAPTOETHANOL, ZINC ION, siah-1A protein | | Authors: | Polekhina, G, House, C.M, Traficante, N, Mackay, J.P, Relaix, F, Sassoon, D.A, Parker, M.W, Bowtell, D.D.L. | | Deposit date: | 2001-09-26 | | Release date: | 2001-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling.
Nat.Struct.Biol., 9, 2002
|
|
1JN7
 
 | |
2KZH
 
 | |
2K1P
 
 | |
2MF8
 
 | | HADDOCK model of MyT1 F4F5 - DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*AP*AP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*AP*AP*CP*TP*TP*TP*CP*GP*GP*T)-3'), Myelin transcription factor 1, ... | | Authors: | Gamsjaeger, R, O'Connell, M.R, Cubeddu, L, Shepherd, N.E, Lowry, J.A, Kwan, A.H, Vandevenne, M, Swanton, M.K, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A structural analysis of DNA binding by myelin transcription factor 1 double zinc fingers.
J.Biol.Chem., 288, 2013
|
|
2JYD
 
 | | Structure of the fifth zinc finger of Myelin Transcription Factor 1 | | Descriptor: | F5 domain of Myelin transcription factor 1, ZINC ION | | Authors: | Gamsjaeger, R, Swanton, M.K, Kobus, F.J, Lehtomaki, E, Lowry, J.A, Kwan, A.H, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2007-12-12 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and biophysical analysis of the DNA binding properties of myelin transcription factor 1.
J.Biol.Chem., 283, 2008
|
|
2JX1
 
 | | Structure of the fifth zinc finger of Myelin Transcription Factor 1 in complex with RARE DNA | | Descriptor: | DNA (5'-D(*DAP*DCP*DCP*DGP*DAP*DAP*DAP*DGP*DTP*DTP*DCP*DAP*DC)-3'), DNA (5'-D(*DGP*DTP*DGP*DAP*DAP*DCP*DTP*DTP*DTP*DCP*DGP*DGP*DT)-3'), Myelin transcription factor 1 | | Authors: | Gamsjaeger, R, Swanton, M.K, Kobus, F.J, Lehtomaki, E, Lowry, J.A, Kwan, A.H, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2007-11-01 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the fifth zinc finger of Myelin Transcription Factor 1 in complex with RARE DNA
To be Published
|
|
2JM3
 
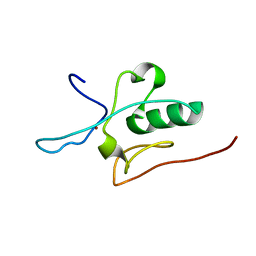 | | Solution structure of the THAP domain from C. elegans C-terminal binding protein (CtBP) | | Descriptor: | Hypothetical protein, ZINC ION | | Authors: | Liew, C.K, Crossley, M, Mackay, J.P, Nicholas, H.R. | | Deposit date: | 2006-09-21 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the THAP domain from Caenorhabditis elegans C-terminal binding protein (CtBP).
J.Mol.Biol., 366, 2007
|
|
2JTN
 
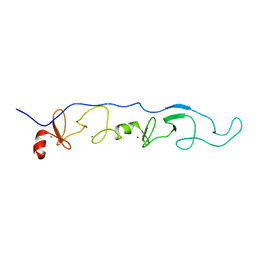 | | NMR Solution Structure of a ldb1-LID:Lhx3-LIM complex | | Descriptor: | LIM domain-binding protein 1, LIM/homeobox protein Lhx3, ZINC ION | | Authors: | Lee, C, Nancarrow, A.L, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2007-08-03 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes
Embo J., 27, 2008
|
|
2N5N
 
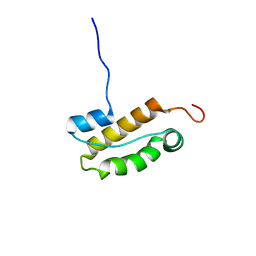 | | Structure of an N-terminal domain of CHD4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Silva, A.P.G, Mackay, J.P. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminal Region of Chromodomain Helicase DNA-binding Protein 4 (CHD4) Is Essential for Activity and Contains a High Mobility Group (HMG) Box-like-domain That Can Bind Poly(ADP-ribose).
J.Biol.Chem., 291, 2016
|
|
2LFN
 
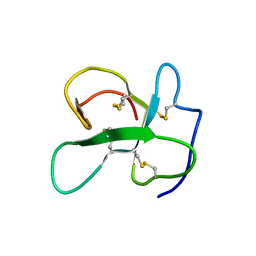 | | Identification of the key regions that drive functional amyloid formation by the fungal hydrophobin EAS | | Descriptor: | Hydrophobin | | Authors: | Macindoe, I, Kwan, A.H, Morris, V.K, Mackay, J.P, Sunde, M. | | Deposit date: | 2011-07-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Self-assembly of functional, amphipathic amyloid monolayers by the fungal hydrophobin EAS
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2L6Z
 
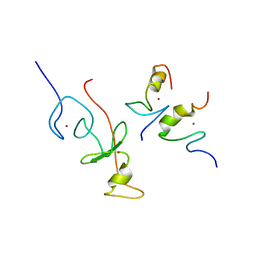 | | haddock model of GATA1NF:Lmo2LIM2-Ldb1LID with FOG | | Descriptor: | Erythroid transcription factor, LIM domain only 2, linker, ... | | Authors: | Wilkinson-White, L, Gamsjaeger, R, Dastmalchi, S, Wienert, B, Stokes, P.H, Crossley, M, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2N39
 
 | |
2MBV
 
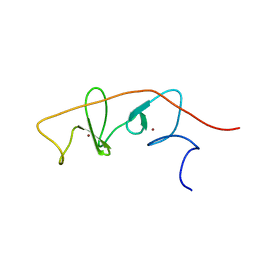 | | LMO4-LIM2 in complex with DEAF1 (404-418) | | Descriptor: | Fusion protein of LIM domain transcription factor LMO4 (77-147) and DEAF1 (404-418), ZINC ION | | Authors: | Joseph, S, Matthews, J.M, Kwan, A.H.-Y, Mackay, J.P, Cubeddu, L, Foo, P. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterisation of LIM-only protein 4 (LMO4) in complex with Deformed Epidermal Autoregulatory Factor-1 (DEAF1)
To be Published
|
|
2KAE
 
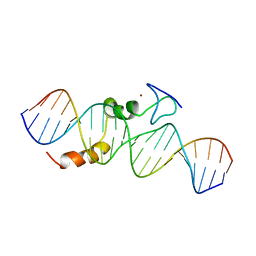 | | data-driven model of MED1:DNA complex | | Descriptor: | 5'-D(*DCP*DGP*DGP*DAP*DAP*DAP*DAP*DGP*DTP*DAP*DTP*DAP*DCP*DTP*DTP*DTP*DTP*DCP*DCP*DG)-3', GATA-type transcription factor, ZINC ION | | Authors: | Lowry, J.A, Gamsjaeger, R, Thong, S, Hung, W, Kwan, A.H, Broitman-Maduro, G, Matthews, J.M, Maduro, M, Mackay, J.P. | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of MED-1 Reveals Unexpected Diversity in the Mechanism of DNA Recognition by GATA-type Zinc Finger Domains.
J.Biol.Chem., 284, 2009
|
|
2L5E
 
 | | Complex between BD1 of Brd3 and GATA-1 C-tail | | Descriptor: | Bromodomain-containing protein 3, GATA-1 | | Authors: | Gamsjaeger, R, Webb, S.R, Lamonica, J.M, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2010-10-31 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3.
Mol. Cell. Biol., 31, 2011
|
|
2L5U
 
 | |
2L6Y
 
 | | haddock model of GATA1NF:Lmo2LIM2-Ldb1LID | | Descriptor: | Erythroid transcription factor, LIM domain only 2, linker, ... | | Authors: | Wilkinson-White, L, Gamsjaeger, R, Dastmalchi, S, Wienert, B, Stokes, P.H, Crossley, M, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2010-12-01 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2L75
 
 | |
5VJJ
 
 | | Crystal structure of the flax-rust effector AvrP | | Descriptor: | Avirulence protein AvrP123, ZINC ION | | Authors: | Zhang, X, Ericsson, D.J, Williams, S.J, Kobe, B. | | Deposit date: | 2017-04-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the Melampsora lini effector AvrP reveals insights into a possible nuclear function and recognition by the flax disease resistance protein P.
Mol. Plant Pathol., 19, 2018
|
|
3SZK
 
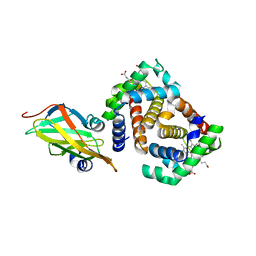 | | Crystal Structure of Human metHaemoglobin Complexed with the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein H, ... | | Authors: | Jacques, D.A, Kumar, K.K, Guss, J.M, Gell, D.A. | | Deposit date: | 2011-07-19 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for hemoglobin capture by Staphylococcus aureus cell-surface protein, IsdH
J.Biol.Chem., 286, 2011
|
|
4FC3
 
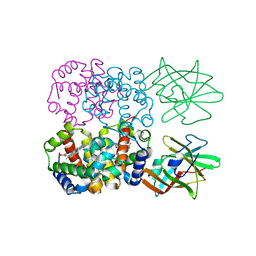 | | Crystal Structure of Human Methaemoglobin Complexed with the Second NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein H, ... | | Authors: | Krishna Kumar, K, Jacques, D.A, Guss, J.M, Gell, D.A. | | Deposit date: | 2012-05-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of the Hemoglobin-IsdH Complex Reveals the Molecular Basis of Iron Capture by Staphylococcus aureus
J.Biol.Chem., 289, 2014
|
|
4IJ2
 
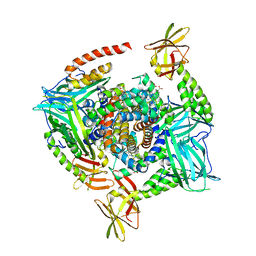 | | Human methemoglobin in complex with the second and third NEAT domains of IsdH from Staphylococcus aureus | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein H, ... | | Authors: | Dickson, C.F, Jacques, D.A, Guss, J.M, Gell, D.A. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.24 Å) | | Cite: | Structure of the Hemoglobin-IsdH Complex Reveals the Molecular Basis of Iron Capture by Staphylococcus aureus
J.Biol.Chem., 289, 2014
|
|
