4Z55
 
 | |
7N3U
 
 | | Crystal structure of human WEE1 kinase domain in complex with ZN-c3 | | Descriptor: | 1-[(7R)-7-ethyl-7-hydroxy-6,7-dihydro-5H-cyclopenta[b]pyridin-2-yl]-6-[4-(4-methylpiperazin-1-yl)anilino]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, Wee1-like protein kinase | | Authors: | Lee, C.C. | | Deposit date: | 2021-06-02 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of ZN-c3, a Highly Potent and Selective Wee1 Inhibitor Undergoing Evaluation in Clinical Trials for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8TZN
 
 | |
9BDI
 
 | |
9BDH
 
 | |
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8ZLR
 
 | | SFX structure of CraCRY in the semiquinone state | | Descriptor: | Cryptochrome photoreceptor, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Maestre-Reyna, M, Hosokawa, Y, Wang, P.-H, Saft, M, Caramello, N, Engilberge, S, Franz-Badur, S, Ngura Putu, E.P.G, Nakamura, M, Wu, W.-J, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Yang, C.-H, Lin, W.-T, Yang, K.-C, Ban, Y, Imura, T, Kazuoka, A, Tanida, E, Owada, S, Joti, Y, Tanaka, R, Tanaka, T, Luo, F, Tono, K, Kiontke, S, Korf, L, Umena, Y, Tosha, T, Bessho, Y, Nango, E, Iwata, S, Royant, A, Tsai, M.-D, Yamamoto, J, Essen, L.-O. | | Deposit date: | 2024-05-21 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Capturing structural intermediates in an animal-like cryptochrome photoreceptor by time-resolved crystallography.
Sci Adv, 11, 2025
|
|
4V0Q
 
 | | Dengue Virus Full Length NS5 Complexed with SAH | | Descriptor: | ACETATE ION, GLYCEROL, NS5 POLYMERASE, ... | | Authors: | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
4V0R
 
 | | DENGUE VIRUS FULL LENGTH NS5 COMPLEXED WITH GTP AND SAH | | Descriptor: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
7F5O
 
 | | Crystal structure of PTPN2 catalytic domain | | Descriptor: | IODIDE ION, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Singh, J.P, Lin, M.-J, Hsu, S.-F, Lee, C.-C, Meng, T.-C. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of TCPTP Unravels an Allosteric Regulatory Role of Helix alpha 7 in Phosphatase Activity.
Biochemistry, 60, 2021
|
|
7F8T
 
 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-07-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
6XNG
 
 | | MHC-like protein complex structure | | Descriptor: | (3R)-N-[(2S,3R)-1-(alpha-D-galactopyranosyloxy)-3-hydroxyheptadecan-2-yl]-3-hydroxyheptadecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Thirunavukkarasu, P, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-07-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Host immunomodulatory lipids created by symbionts from dietary amino acids.
Nature, 600, 2021
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YC7
 
 | | Dark, fully reduced structure of the MmCPDII-DNA complex as produced at SwissFEL | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YD7
 
 | | TR-SFX MmCPDII-DNA complex: 1 ns snapshot. Includes 1 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCM
 
 | | TR-SFX MmCPDII-DNA complex: 100 ps snapshot. Includes 100ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCR
 
 | | TR-SFX MmCPDII-DNA complex: 450 ps snapshot. Includes 450ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YCP
 
 | | TR-SFX MmCPDII-DNA complex: 250 ps snapshot. Includes 250 ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YD8
 
 | | TR-SFX MmCPDII-DNA complex: 2 ns snapshot. Includes 2 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YD6
 
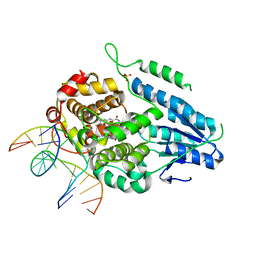 | | TR-SFX MmCPDII-DNA complex: 650 ps snapshot. Includes 650ps, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YDZ
 
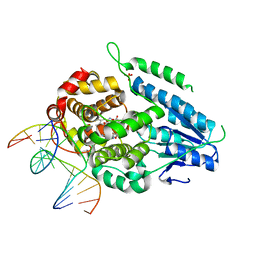 | | TR-SFX MmCPDII-DNA complex: dark state as collected in SACLA | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YE0
 
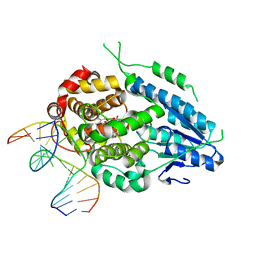 | | DF-SFX MmCPDII-DNA complex: steady state oxidized complex | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEE
 
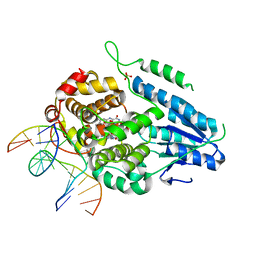 | | TR-SFX MmCPDII-DNA complex: 10 ns snapshot. Includes 10 ns, dark, and extrapolated structure factors. Collected at SwissFEL | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
