6R69
 
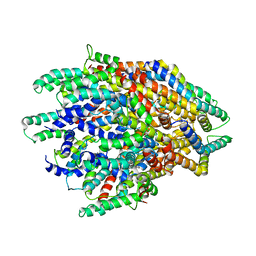 | | Improved map of the FliPQR complex that forms the core of the Salmonella type III secretion system export apparatus. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Johnson, S, Kuhlen, L, Abrusci, P, Lea, S.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | The Structure of an Injectisome Export Gate Demonstrates Conservation of Architecture in the Core Export Gate between Flagellar and Virulence Type III Secretion Systems.
Mbio, 10, 2019
|
|
8SA2
 
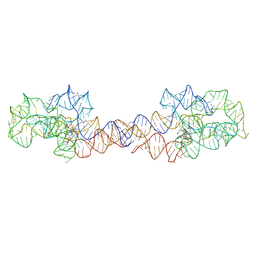 | | Adenosylcobalamin-bound riboswitch dimer, form 1 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 1 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA3
 
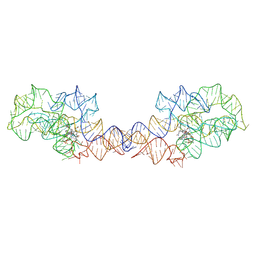 | | Adenosylcobalamin-bound riboswitch dimer, form 2 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 2 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA6
 
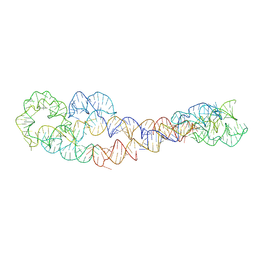 | | apo form of adenosylcobalamin riboswitch dimer | | Descriptor: | apo form of adenosylcobalamin riboswitch dimer | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
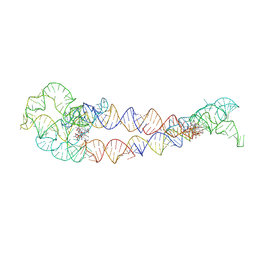 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA4
 
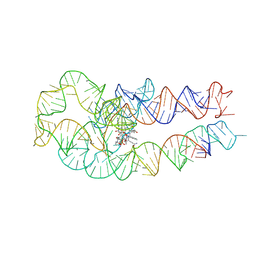 | | Adenosylcobalamin-bound riboswitch dimer, form 3 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 3 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7SB2
 
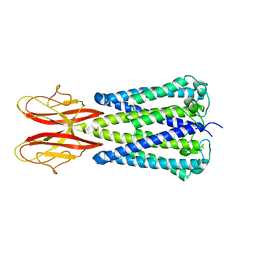 | |
7SAT
 
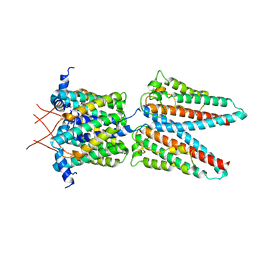 | |
7SAZ
 
 | |
7SAU
 
 | |
7SAX
 
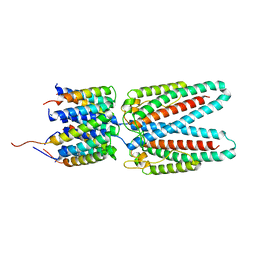 | |
6RQJ
 
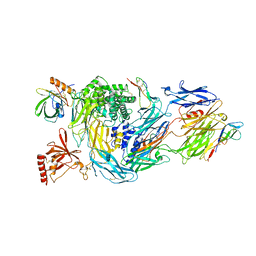 | | Structure of human complement C5 complexed with tick inhibitors OmCI, RaCI1 and CirpT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5, Complement inhibitor, ... | | Authors: | Reichhardt, M.P, Johnson, S, Lea, S.M. | | Deposit date: | 2019-05-15 | | Release date: | 2020-01-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An inhibitor of complement C5 provides structural insights into activation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5MWE
 
 | | Complex between the Leucine Zipper (LZ, residues 490-567) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) | | Descriptor: | 1,2-ETHANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, Centrosomin, ... | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
5MW9
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) - L535E mutant form | | Descriptor: | Centrosomin, ZINC ION | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
5MVW
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Centrosomin, ... | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
5MW0
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) - L535E mutant form | | Descriptor: | Centrosomin, ZINC ION | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
6RPT
 
 | |
5MWF
 
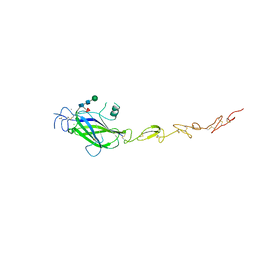 | | Human Jagged2 C2-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MVX
 
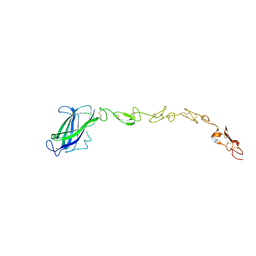 | | Human DLL4 C2-EGF3 | | Descriptor: | Delta-like protein 4, alpha-L-fucopyranose | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MW7
 
 | | Human Jagged2 C2-EGF3 | | Descriptor: | Protein jagged-2, alpha-L-fucopyranose | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MWB
 
 | | Human Notch-2 EGF11-13 | | Descriptor: | CALCIUM ION, Neurogenic locus notch homolog protein 2, alpha-L-fucopyranose, ... | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
5MW5
 
 | | Human Jagged2 C2-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protein jagged-2 | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
6S3L
 
 | | Structure of the core of the flagellar export apparatus from Vibrio mimicus, the FliPQR-FlhB complex. | | Descriptor: | Flagellar biosynthetic protein FlhB, Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, ... | | Authors: | Kuhlen, L, Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2019-06-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The substrate specificity switch FlhB assembles onto the export gate to regulate type three secretion.
Nat Commun, 11, 2020
|
|
6S3R
 
 | | Structure of the FliPQR complex from the flagellar type 3 secretion system of Pseudomonas savastanoi. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Kuhlen, L, Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2019-06-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The substrate specificity switch FlhB assembles onto the export gate to regulate type three secretion.
Nat Commun, 11, 2020
|
|
6SD1
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 33-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
