5FMV
 
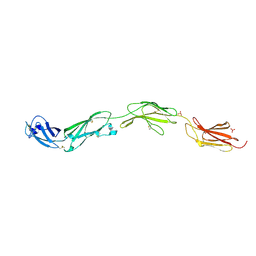 | | Crystal structure of human CD45 extracellular region, domains d1-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C, SULFATE ION | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FN6
 
 | | Crystal structure of human CD45 extracellular region, domains d1-d3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FN8
 
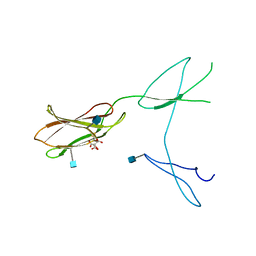 | | Crystal structure of rat CD45 extracellular region, domains d3-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FYN
 
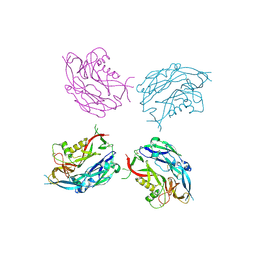 | | Sub-tomogram averaging of Tula virus glycoprotein spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PUUMALA VIRUS GN GLYCOPROTEIN, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, S, Rissanen, I, Zeltina, A, Hepojoki, J, Raghwani, J, Harlos, K, Pybus, O.G, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (15.6 Å) | | Cite: | A Molecular-Level Account of the Antigenic Hantaviral Surface.
Cell Rep., 15, 2016
|
|
6JRE
 
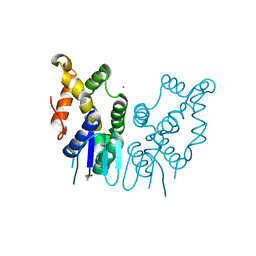 | | Structure of N-terminal domain of Plasmodium vivax p43 (PfNTD) solved by Co-SAD phasing | | Descriptor: | Aminoacyl-tRNA synthetase-interacting multifunctional protein p43, COBALT (II) ION | | Authors: | Manickam, Y, Harlos, K, Sharma, M, Gupta, S, Sharma, A. | | Deposit date: | 2019-04-03 | | Release date: | 2020-03-11 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2JJS
 
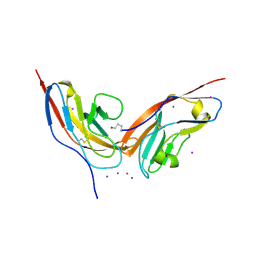 | | Structure of human CD47 in complex with human signal regulatory protein (SIRP) alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, LEUKOCYTE SURFACE ANTIGEN CD47, ... | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47.
Mol. Cell, 31, 2008
|
|
1QFT
 
 | | HISTAMINE BINDING PROTEIN FROM FEMALE BROWN EAR RHIPICEPHALUS APPENDICULATUS | | Descriptor: | HISTAMINE, PROTEIN (FEMALE-SPECIFIC HISTAMINE BINDING PROTEIN 2) | | Authors: | Paesen, G.C, Adams, P.L, Harlos, K, Nuttal, P.A, Stuart, D.I. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tick histamine-binding proteins: isolation, cloning, and three-dimensional structure.
Mol.Cell, 3, 1999
|
|
1QFV
 
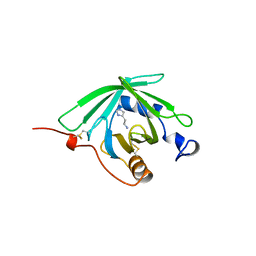 | | HISTAMINE BINDING PROTEIN FROM FEMALE BROWN EAR RHIPICEPHALUS APPENDICULATUS | | Descriptor: | HISTAMINE, PROTEIN (FEMALE-SPECIFIC HISTAMINE BINDING PROTEIN 2) | | Authors: | Paesen, G.C, Adams, P.L, Harlos, K, Nuttal, P.A, Stuart, D.I. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Tick histamine-binding proteins: isolation, cloning, and three-dimensional structure.
Mol.Cell, 3, 1999
|
|
5ZH3
 
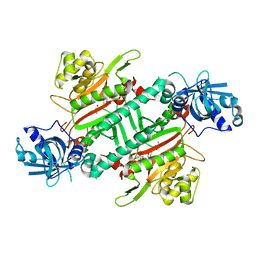 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-6 | | Descriptor: | (3S)-6,8-dihydroxy-3-{[(2R,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, LYSINE, Lysine-tRNA ligase | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
5ZH5
 
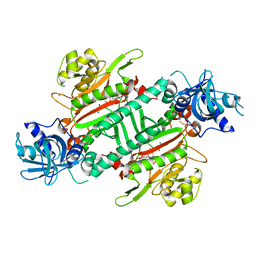 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-2 | | Descriptor: | (3S)-6,8-dihydroxy-3-{[(2R,6S)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, CHLORIDE ION, LYSINE, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
5ZKF
 
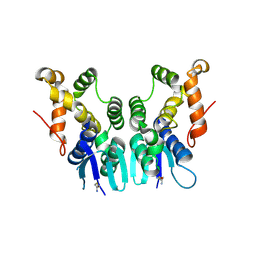 | | Crystal Structure of N-terminal Domain of Plasmodium vivax p43 in space group P21 | | Descriptor: | Aminoacyl tRNA Synthetase Complex-Interacting Multifunctional Protein p43 | | Authors: | Gupta, S, Sharma, M, Harlos, K, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-24 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5ZH2
 
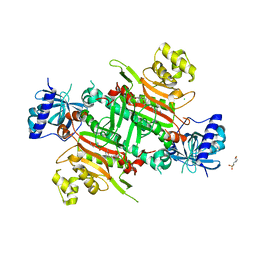 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-5 | | Descriptor: | (3R)-6,8-dihydroxy-3-{[(2R,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
5ZKE
 
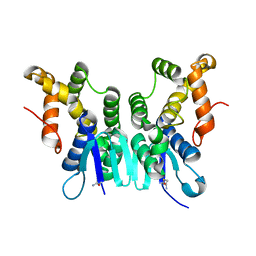 | | Crystal Structure of N-terminal Domain of Plasmodium vivax p43 in space group P212121 | | Descriptor: | Aminoacyl tRNA Synthetase Complex-Interacting Multifunctional Protein p43 | | Authors: | Gupta, S, Sharma, M, Harlos, K, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5ZH4
 
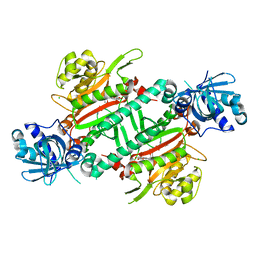 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-7 | | Descriptor: | (3R)-6,8-dihydroxy-3-{[(2S,6R)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, CHLORIDE ION, LYSINE, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
1HNF
 
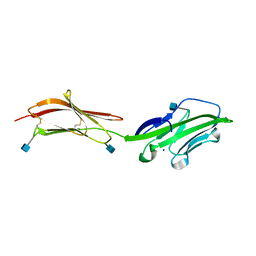 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR REGION OF THE HUMAN CELL ADHESION MOLECULE CD2 AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD2, SODIUM ION | | Authors: | Bodian, D.L, Jones, E.Y, Harlos, K, Stuart, D.I, Davis, S.J. | | Deposit date: | 1994-08-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 A resolution.
Structure, 2, 1994
|
|
5NUZ
 
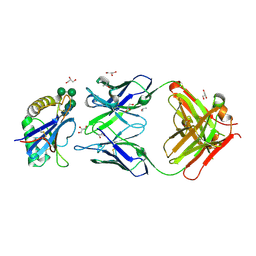 | | Junin virus GP1 glycoprotein in complex with an antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Zeltina, A, Krumm, S.A, Sahin, M, Struwe, W.B, Harlos, K, Nunberg, J.H, Crispin, M, Pinschewer, D.D, Doores, K.J, Bowden, T.A. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Convergent immunological solutions to Argentine hemorrhagic fever virus neutralization.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6GGM
 
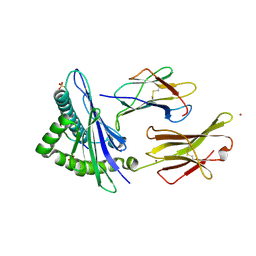 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P2-Phe. | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Mtb44*P2-Phe peptide variant (ARG-PHE-PRO-ALA-LYS-ALA-PRO-LEU-LEU), ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
6GH4
 
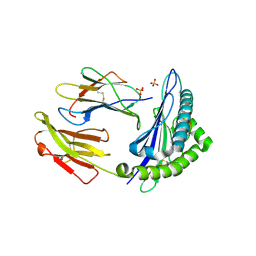 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P2-Gln. | | Descriptor: | ARG-GLN-PRO-ALA-LYS-ALA-PRO-LEU-LEU, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
6GH1
 
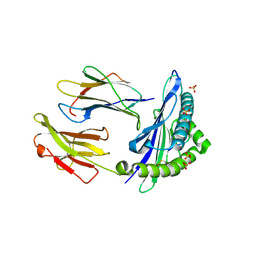 | | HLA-E*01:03 in complex with Mtb44 | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], MHC class I antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
1GVG
 
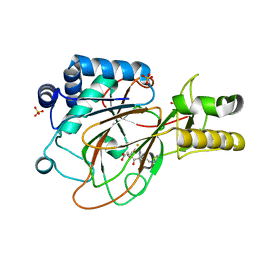 | | Crystal Structure of Clavaminate Synthase with Nitric Oxide | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, DEOXYGUANIDINOPROCLAVAMINIC ACID, ... | | Authors: | Zhang, Z.H, Ren, J, McKinnon, C.H, Clifton, I.J, Harlos, K, Schofield, C.J. | | Deposit date: | 2002-02-12 | | Release date: | 2003-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of a Clavaminate Synthase-Fe(II) -2-Oxoglutarate-Substrate-No Complex: Evidence for Metal Centered Rearrangements
FEBS Lett., 517, 2002
|
|
5OPG
 
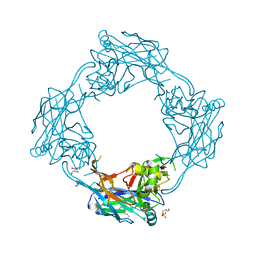 | | Structure of the Hantaan virus Gn glycoprotein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Rissanen, I, Stass, R, Zeltina, A, Li, S, Hepojoki, J, Harlos, K, Gilbert, R.J.C, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2017-08-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Transitions of the Conserved and Metastable Hantaviral Glycoprotein Envelope.
J. Virol., 91, 2017
|
|
6GHN
 
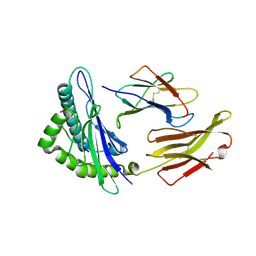 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P9-Phe. | | Descriptor: | ARG-LEU-PRO-ALA-LYS-ALA-PRO-LEU-PHE, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
5MGS
 
 | | Human receptor NKR-P1 in deglycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1 | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
5OSN
 
 | | Crystal Structure of Bovine Enterovirus 2 determined with Serial Femtosecond X-ray Crystallography | | Descriptor: | Capsid protein, GLUTAMIC ACID, POTASSIUM ION, ... | | Authors: | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Kotecha, A, Kelly, J, Rowlands, D.J, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
6GL1
 
 | | HLA-E*01:03 in complex with the HIV epitope, RL9HIV | | Descriptor: | ARG-MET-TYR-SER-PRO-THR-SER-ILE-LEU, Beta-2-microglobulin, MHC class I antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
