6C69
 
 | | Structure of glycolipid aGSA[12,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6C6E
 
 | | Structure of glycolipid aGSA[26,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6CWB
 
 | | Structure of alpha-GSA[8,4P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-[(1E)-4-phenylbutylidene]-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}octanamide (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6CX5
 
 | | Structure of alpha-GSA[8,8P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(8-phenyloctyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}octanamide (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6CX9
 
 | | Structure of alpha-GSA[16,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6C6A
 
 | | Structure of glycolipid aGSA[16,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6CW9
 
 | | Structure of alpha-GC[8,16P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6CX7
 
 | | Structure of alpha-GSA[12,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-04-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
4F7E
 
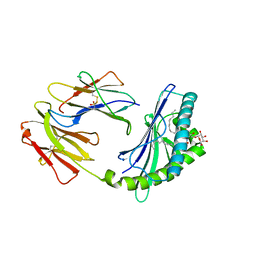 | | Crystal structure of bovine CD1d with bound C16:0-alpha-galactosyl ceramide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Wang, J, Zajonc, D.M. | | Deposit date: | 2012-05-15 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Bovine CD1d Reveal Altered αGalCer Presentation and a Restricted A' Pocket Unable to Bind Long-Chain Glycolipids.
Plos One, 7, 2012
|
|
4F7C
 
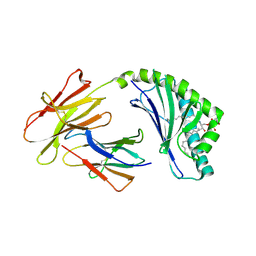 | | Crystal structure of bovine CD1d with bound C12-di-sulfatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CD1D antigen, ... | | Authors: | Wang, J, Zajonc, D.M. | | Deposit date: | 2012-05-15 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | Crystal Structures of Bovine CD1d Reveal Altered αGalCer Presentation and a Restricted A' Pocket Unable to Bind Long-Chain Glycolipids.
Plos One, 7, 2012
|
|
1OJR
 
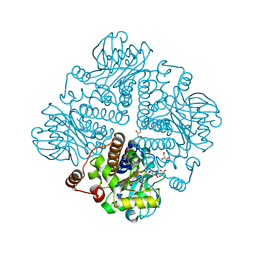 | | L-rhamnulose-1-phosphate aldolase from Escherichia coli (mutant E192A) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Dihydroxyacetone, GLYCEROL, ... | | Authors: | Kroemer, M, Merkel, I, Schulz, G.E. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Catalytic Mechanism of L-Rhamnulose-1-Phosphate Aldolase.
Biochemistry, 42, 2003
|
|
5OCD
 
 | |
1GT7
 
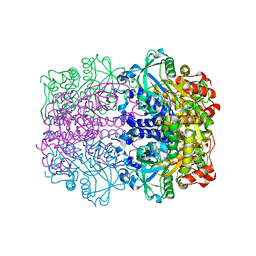 | | L-rhamnulose-1-phosphate aldolase from Escherichia coli | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, RHAMNULOSE-1-PHOSPHATE ALDOLASE, ZINC ION | | Authors: | Kroemer, M, Schulz, G.E. | | Deposit date: | 2002-01-14 | | Release date: | 2002-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of L-Rhamnulose-1-Phosphate Aldolase (Class II) Solved by Low-Resolution Sir Phasing and 20-Fold Ncs Averaging
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6Y4B
 
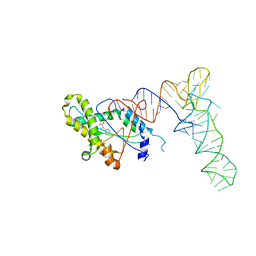 | |
6Y3G
 
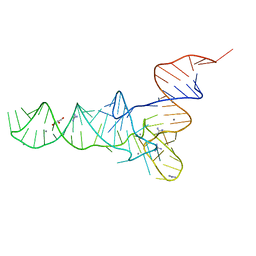 | | Crystal structure of phenylalanine tRNA from Escherichia coli | | Descriptor: | CALCIUM ION, GLYCEROL, GUANIDINE, ... | | Authors: | Bourgeois, G, Mechulam, Y, Schmitt, E. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the interaction between cyclodipeptide synthases and aminoacylated tRNA substrates.
Rna, 26, 2020
|
|
4FUA
 
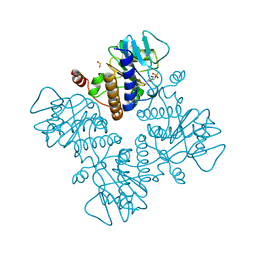 | | L-FUCULOSE-1-PHOSPHATE ALDOLASE COMPLEX WITH PGH | | Descriptor: | BETA-MERCAPTOETHANOL, L-FUCULOSE-1-PHOSPHATE ALDOLASE, PHOSPHOGLYCOLOHYDROXAMIC ACID, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Catalytic mechanism of the metal-dependent fuculose aldolase from Escherichia coli as derived from the structure.
J.Mol.Biol., 259, 1996
|
|
7MT4
 
 | | Crystal structure of tryptophan Synthase in complex with F9, NH4+, pH7.8 - alpha aminoacrylate form - E(A-A) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, AMMONIUM ION, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MT5
 
 | | Crystal structure of tryptophan synthase in complex with F9, Cs+, pH7.8 - alpha aminoacrylate form - E(A-A) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, CESIUM ION, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MT6
 
 | | Crystal structure of tryptophan synthase in complex with F9, Cs+, benzimidazole, pH7.8 - alpha aminoacrylate form - E(A-A)(BZI) | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Drago, V, Hilario, E, Dunn, M.F, Mueser, T.C, Mueller, L.J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Imaging active site chemistry and protonation states: NMR crystallography of the tryptophan synthase alpha-aminoacrylate intermediate.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3S6C
 
 | | Structure of human CD1e | | Descriptor: | Beta-2-microglobulin, T-cell surface glycoprotein CD1e, membrane-associated, ... | | Authors: | Garcia-Alles, L.F, Maveyraud, L, Tranier, S, Mourey, L. | | Deposit date: | 2011-05-25 | | Release date: | 2011-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3T8X
 
 | | Crystal structure of human CD1b in complex with synthetic antigenic diacylsulfoglycolipid SGL12 and endogenous spacer | | Descriptor: | 2-O-sulfo-alpha-D-glucopyranosyl 2-O-hexadecanoyl-3-O-[(2E,4S,6S,8S)-2,4,6,8-tetramethyltetracos-2-enoyl]-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Garcia-Alles, L.F, Maveyraud, L, Mourey, L, Julien, S. | | Deposit date: | 2011-08-02 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural reorganization of the human CD1b Antigen-binding groove for presentation of mycobacterial sulfoglycolipids
To be Published
|
|
7M6G
 
 | |
7M6I
 
 | |
7M6E
 
 | |
7M6H
 
 | |
