5BTP
 
 | | Fusobacterium ulcerans ZTP riboswitch bound to ZMP | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.816 Å) | | Cite: | Recognition of the bacterial alarmone ZMP through long-distance association of two RNA subdomains.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5BJO
 
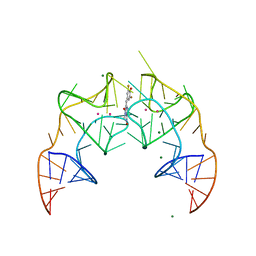 | | Crystal structure of the Corn RNA aptamer in complex with DFHO, site-specific 5-iodo-U | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Warner, K.D, Song, W, Filonov, G.S, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2016-08-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A homodimer interface without base pairs in an RNA mimic of red fluorescent protein.
Nat. Chem. Biol., 13, 2017
|
|
5BJP
 
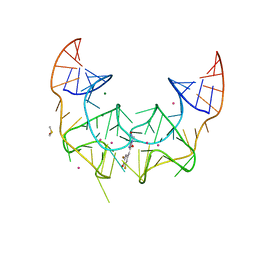 | | Crystal structure of the Corn RNA aptamer in complex with DFHO, iridium hexammine soak | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, DIMETHYL SULFOXIDE, IRIDIUM ION, ... | | Authors: | Warner, K.D, Song, W, Filonov, G.S, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2016-08-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | A homodimer interface without base pairs in an RNA mimic of red fluorescent protein.
Nat. Chem. Biol., 13, 2017
|
|
6C64
 
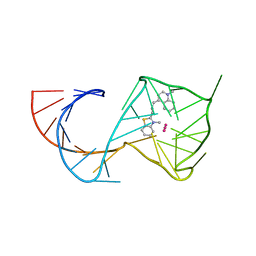 | | Crystal Structure of the Mango-II Fluorescent Aptamer Bound to TO3-Biotin | | Descriptor: | 1-methyl-4-[(1E)-3-(3-methyl-1,3-benzothiazol-3-ium-2-yl)prop-1-en-1-yl]quinolin-1-ium, POTASSIUM ION, RNA (32-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.00014877 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
4MEH
 
 | |
4NYC
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thieno[2,3-b]pyrazin-7-amine | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, thiM TPP riboswitch, ... | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
6C65
 
 | | Crystal Structure of the Mango-II-A22U Fluorescent Aptamer Bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, POTASSIUM ION, RNA (35-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.80005169 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
4NYD
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with hypoxanthine | | Descriptor: | HYPOXANTHINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
4NYA
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with 5-(azidomethyl)-2-methylpyrimidin-4-amine | | Descriptor: | 5-(azidomethyl)-2-methylpyrimidin-4-amine, MAGNESIUM ION, thiM TPP riboswitch | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
6C63
 
 | | Crystal Structure of the Mango-II Fluorescent Aptamer Bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, POTASSIUM ION, RNA (32-MER), ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-01-17 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.900028 Å) | | Cite: | Crystal Structures of the Mango-II RNA Aptamer Reveal Heterogeneous Fluorophore Binding and Guide Engineering of Variants with Improved Selectivity and Brightness.
Biochemistry, 57, 2018
|
|
6CB3
 
 | |
6CC3
 
 | |
4NYB
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with (4-(1,2,3-thiadiazol-4-yl)phenyl)methanamine | | Descriptor: | 1-[4-(1,2,3-thiadiazol-4-yl)phenyl]methanamine, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
4NYG
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thiamine | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, MAGNESIUM ION, thiM TPP riboswitch | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
6CC1
 
 | |
1SB7
 
 | |
6E8T
 
 | |
6E1W
 
 | | Crystal structure of a class I PreQ1 riboswitch complexed with PreQ1 | | Descriptor: | 2-amino-5-(aminomethyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1S
 
 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 1: 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine | | Descriptor: | 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1T
 
 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 1: 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine, MAGNESIUM ION, ... | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1U
 
 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 2: 2-[(dibenzo[b,d]furan-2-yl)oxy]-N,N-dimethylethan-1-amine | | Descriptor: | 2-[(dibenzo[b,d]furan-2-yl)oxy]-N,N-dimethylethan-1-amine, MAGNESIUM ION, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E8U
 
 | | Structure of the Mango-III (A10U) aptamer bound to TO1-Biotin | | Descriptor: | 4-[(3-{2,16-dioxo-20-[(3aR,4R,6aS)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and functional reselection of the Mango-III fluorogenic RNA aptamer.
Nat. Chem. Biol., 15, 2019
|
|
6E1V
 
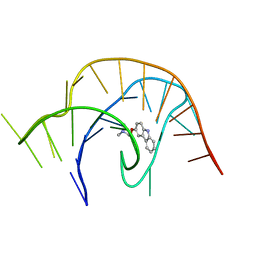 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 3: 2-[(9H-carbazol-3-yl)oxy]-N,N-dimethylethan-1-amine | | Descriptor: | 2-[(9H-carbazol-3-yl)oxy]-N,N-dimethylethan-1-amine, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E8S
 
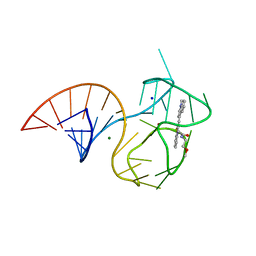 | | Structure of the iMango-III aptamer bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and functional reselection of the Mango-III fluorogenic RNA aptamer.
Nat. Chem. Biol., 15, 2019
|
|
6E82
 
 | |
