5N7E
 
 | | Crystal structure of the Dbl-homology domain of Bcr-Abl in complex with monobody Mb(Bcr-DH_4). | | Descriptor: | Breakpoint cluster region protein, Mb(Bcr-DH_4) | | Authors: | Reckel, S, Reynaud, A, Pojer, F, Hantschel, O. | | Deposit date: | 2017-02-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
6F9A
 
 | | Solution structure of the MRH domain of Yos9 complexed with alpha3,alpha6-Man5 | | Descriptor: | ER quality-control lectin | | Authors: | Kniss, A, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MRH domain of Yos9 complexed with alpha3,alpha6-Man5
To Be Published
|
|
6RYB
 
 | |
6RYA
 
 | | Structure of Dup1 mutant H67A:Ubiquitin complex | | Descriptor: | Polyubiquitin-C, Septation initiation protein | | Authors: | Donghyuk, S, Ivan, D. | | Deposit date: | 2019-06-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Regulation of Phosphoribosyl-Linked Serine Ubiquitination by Deubiquitinases DupA and DupB.
Mol.Cell, 77, 2020
|
|
8CHP
 
 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfinyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfinyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHR
 
 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((R)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHN
 
 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHI
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-3,5-dichloro-N-methylphenylsulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[S-[3,5-bis(chloranyl)phenyl]-N-methyl-sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHK
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHL
 
 | | Human FKBP12 in complex with (1S,5S,6R)-9-((3,5-dichlorophenyl)sulfonyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,9-diazabicyclo[4.2.1]nonan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, CADMIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHQ
 
 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((S)-3,5-dichloro-N-methylphenylsulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[S-[3,5-bis(chloranyl)phenyl]-N-methyl-sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHM
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfinyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfinyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHJ
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((R)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
2KBY
 
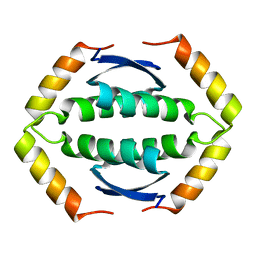 | | The Tetramerization Domain of Human p73 | | Descriptor: | Tumor protein p73 | | Authors: | Coutandin, D, Ikeya, T, Loehr, F, Guntert, P, Ou, H.D, Doetsch, V. | | Deposit date: | 2008-12-12 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conformational stability and activity of p73 require a second helix in the tetramerization domain.
Cell Death Differ., 16, 2009
|
|
2KX7
 
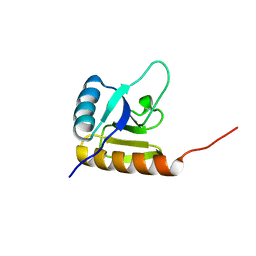 | | Solution structure of the E.coli RcsD-ABL domain (residues 688-795) | | Descriptor: | Sensor-like histidine kinase yojN | | Authors: | Rogov, V.V, Schmoee, K, Rogova, N.Y, Loehr, F, Bernhard, F, Doetsch, V. | | Deposit date: | 2010-04-27 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Rcs Phosphotransfer: The Newly Identified RcsD-ABL Domain Enhances Interaction with the Response Regulator RcsB.
Structure, 19, 2011
|
|
1QA9
 
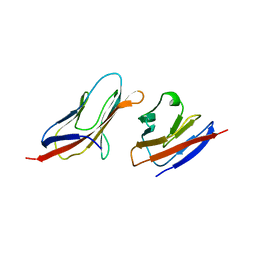 | | Structure of a Heterophilic Adhesion Complex Between the Human CD2 and CD58(LFA-3) Counter-Receptors | | Descriptor: | HUMAN CD2 PROTEIN, HUMAN CD58 PROTEIN | | Authors: | Wang, J.-H, Smolyar, A, Tan, K, Liu, J.-H, Kim, M, Sun, Z.J, Wagner, G, Reinherz, E.L. | | Deposit date: | 1999-04-13 | | Release date: | 1999-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors.
Cell(Cambridge,Mass.), 97, 1999
|
|
2L8Y
 
 | | Solution structure of the E. coli outer membrane protein RcsF (periplasmatic domain) | | Descriptor: | Protein rcsF | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Lohr, F, Doetsch, V. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RCSF.
J.Biol.Chem., 286, 2011
|
|
2L8J
 
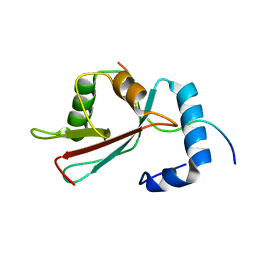 | | GABARAPL-1 NBR1-LIR complex structure | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, NBR1-LIR peptide | | Authors: | Rogov, V.V, Rozenknop, A, Rogova, N.Y, Loehr, F, Guentert, P, Dikic, I, Doetsch, V. | | Deposit date: | 2011-01-17 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Interaction of GABARAPL-1 with the LIR Motif of NBR1.
J.Mol.Biol., 410, 2011
|
|
2M8J
 
 | | Structure of Pin1 WW domain phospho-mimic S16E | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
2M8I
 
 | | Structure of Pin1 WW domain | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
2MD9
 
 | | Solution Structure of an Active Site Mutant Pepitdyl Carrier Protein | | Descriptor: | Tyrocidine synthase 3 | | Authors: | Tufar, P, Rahighi, S, Kraas, F.I, Kirchner, D.K, Loehr, F, Henrich, E, Koepke, J, Dikic, I, Guentert, P, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal Structure of a PCP/Sfp Complex Reveals the Structural Basis for Carrier Protein Posttranslational Modification.
Chem.Biol., 21, 2014
|
|
2MYX
 
 | | Structure of the CUE domain of yeast Cue1 | | Descriptor: | Coupling of ubiquitin conjugation to ER degradation protein 1 | | Authors: | Kniss, A, Rogov, V.V, Loehr, F, Guentert, P, Doetsch, V. | | Deposit date: | 2015-02-02 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The CUE Domain of Cue1 Aligns Growing Ubiquitin Chains with Ubc7 for Rapid Elongation.
Mol.Cell, 62, 2016
|
|
2NB1
 
 | | P63/p73 hetero-tetramerisation domain | | Descriptor: | Tumor protein 63, Tumor protein p73 | | Authors: | Gebel, J, Buchner, L, Loehr, F.M, Luh, L.M, Coutandin, D, Guentert, P, Doetsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
2MW4
 
 | | Tetramerization domain of the Ciona intestinalis p53/p73-b transcription factor protein | | Descriptor: | Transcription factor protein | | Authors: | Heering, J.P, Jonker, H.R.A, Loehr, F, Schwalbe, H, Doetsch, V. | | Deposit date: | 2014-10-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigations of the p53/p73 homologs from the tunicate species Ciona intestinalis reveal the sequence requirements for the formation of a tetramerization domain.
Protein Sci., 25, 2016
|
|
2L6X
 
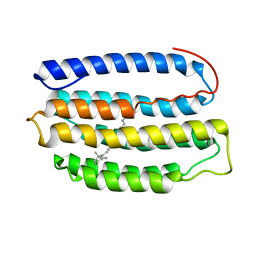 | | Solution NMR Structure of Proteorhodopsin. | | Descriptor: | Green-light absorbing proteorhodopsin, RETINAL | | Authors: | Reckel, S, Gottstein, D, Stehle, J, Loehr, F, Takeda, M, Silvers, R, Kainosho, M, Glaubitz, C, Bernhard, F, Schwalbe, H, Guntert, P, Doetsch, V, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2010-11-29 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of proteorhodopsin.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
