7EK0
 
 | | Complex Structure of antibody BD-503 and RBD-N501Y of COVID-19 | | Descriptor: | Heavy Chain of BD-503, Light Chain of BD-503, Spike protein S1 | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJZ
 
 | | Complex Structure of antibody BD-503 and RBD-S477N of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJY
 
 | | Complex Structure of antibody BD-503 and RBD of COVID-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | Authors: | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
8XMP
 
 | | Structure of CD163 in complex with Hb-Hp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xu, H, Su, X.D. | | Deposit date: | 2023-12-27 | | Release date: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure of CD163 in complex with Hb-Hp
To Be Published
|
|
8XMW
 
 | | Local refinement of Hb-Hp bind to CD163 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xu, H, Su, X.D. | | Deposit date: | 2023-12-28 | | Release date: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Local refinement of Hb-Hp bind to CD163
To Be Published
|
|
8XMK
 
 | | Local refinement of SRCR5-9 domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Scavenger receptor cysteine-rich type 1 protein M130 | | Authors: | Xu, H, Su, X.D. | | Deposit date: | 2023-12-27 | | Release date: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Local refinement of SRCR5-9 domains
To Be Published
|
|
8XMQ
 
 | | Structure of dimeric CD163 in complex with Hb-Hp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xu, H, Su, X.D. | | Deposit date: | 2023-12-27 | | Release date: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure of dimeric CD163 in complex with Hb-Hp
To Be Published
|
|
1BIH
 
 | | CRYSTAL STRUCTURE OF THE INSECT IMMUNE PROTEIN HEMOLIN: A NEW DOMAIN ARRANGEMENT WITH IMPLICATIONS FOR HOMOPHILIC ADHESION | | Descriptor: | HEMOLIN, PHOSPHATE ION | | Authors: | Su, X.-D, Gastinel, L.N, Vaughn, D.E, Faye, I, Poon, P, Bjorkman, P.J. | | Deposit date: | 1998-06-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of hemolin: a horseshoe shape with implications for homophilic adhesion.
Science, 281, 1998
|
|
1PHR
 
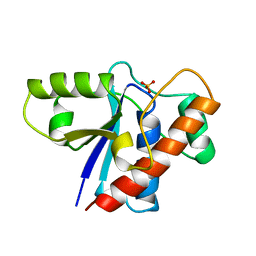 | | THE CRYSTAL STRUCTURE OF A LOW MOLECULAR PHOSPHOTYROSINE PROTEIN PHOSPHATASE | | Descriptor: | LOW MOLECULAR WEIGHT PHOSPHOTYROSINE PROTEIN PHOSPHATASE, SULFATE ION | | Authors: | Su, X.-D, Taddei, N, Stefani, M, Ramponi, G, Nordlund, P. | | Deposit date: | 1994-07-05 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a low-molecular-weight phosphotyrosine protein phosphatase.
Nature, 370, 1994
|
|
5XNL
 
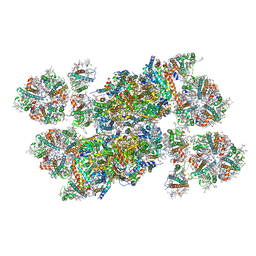 | | Structure of stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNM
 
 | | Structure of unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNN
 
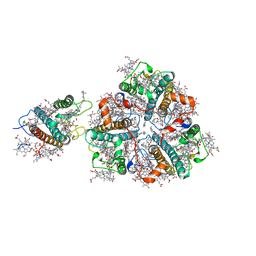 | | Structure of M-LHCII and CP24 complexes in the stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNO
 
 | | Structure of M-LHCII and CP24 complexes in the unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
4L9A
 
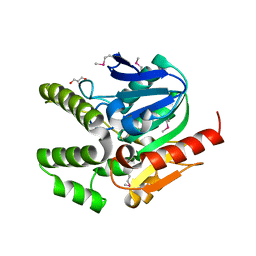 | |
1XIK
 
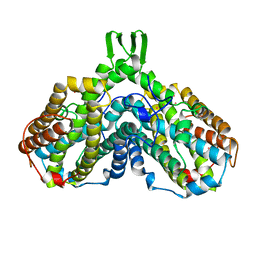 | | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 BETA CHAIN | | Descriptor: | FE (II) ION, MERCURY (II) ION, PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE | | Authors: | Logan, D.T, Su, X.-D, Aberg, A, Regnstrom, K, Hajdu, J, Eklund, H, Nordlund, P. | | Deposit date: | 1996-08-06 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reduced protein R2 of ribonucleotide reductase: the structural basis for oxygen activation at a dinuclear iron site.
Structure, 4, 1996
|
|
1LVA
 
 | |
6L4S
 
 | |
6LRQ
 
 | |
1JL3
 
 | | Crystal Structure of B. subtilis ArsC | | Descriptor: | ARSENATE REDUCTASE, SULFATE ION | | Authors: | Su, X.-D, Bennett, M.S. | | Deposit date: | 2001-07-15 | | Release date: | 2001-10-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
9LNG
 
 | | An antibody target the fusion protein of Nipah virus | | Descriptor: | Fab NiF03-3C9 heavy chain, Fab NiF03-3C9 light chain, Fusion glycoprotein F0 | | Authors: | Xu, H, Su, X.D. | | Deposit date: | 2025-01-21 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | A monoclonal antibody targeting conserved regions of pre-fusion protein cross-neutralizes Nipah and Hendra virus variants.
Antiviral Res., 240, 2025
|
|
3RNY
 
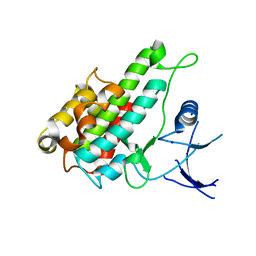 | | Crystal structure of human RSK1 C-terminal kinase domain | | Descriptor: | Ribosomal protein S6 kinase alpha-1, SODIUM ION | | Authors: | Li, D, Fu, T.-M, Nan, J, Su, X.-D. | | Deposit date: | 2011-04-24 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the autoinhibition of the C-terminal kinase domain of human RSK1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4H10
 
 | |
4IYR
 
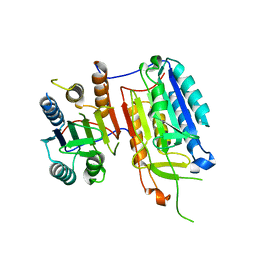 | | Crystal structure of full-length caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Cao, Q, Wang, X.-J, Li, L.-F, Su, X.-D. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | The regulatory mechanism of the caspase 6 pro-domain revealed by crystal structure and biochemical assays.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5GNJ
 
 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*AP*CP*AP*CP*GP*TP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*TP*CP*AP*CP*GP*TP*GP*TP*TP*CP*C)-3'), Transcription factor MYC2 | | Authors: | Lian, T, Xu, Y, Su, X. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Tetrameric Arabidopsis MYC2 Reveals the Mechanism of Enhanced Interaction with DNA.
Cell Rep, 19, 2017
|
|
3CP7
 
 | |
