1RAA
 
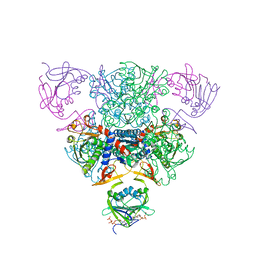 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1OHF
 
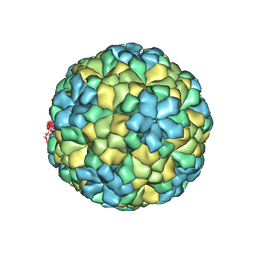 | | The refined structure of Nudaurelia capensis omega virus | | Descriptor: | MAGNESIUM ION, p70 | | Authors: | Helgstrand, C, Munshi, S, Johnson, J.E, Liljas, L. | | Deposit date: | 2003-05-26 | | Release date: | 2004-02-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Refined Structure of Nudaurelia Capensis Omega Virus Reveals Control Elements for a T = 4 Capsid Maturation
Virology, 318, 2004
|
|
1R5T
 
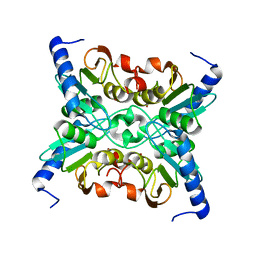 | | The Crystal Structure of Cytidine Deaminase CDD1, an Orphan C to U editase from Yeast | | Descriptor: | Cytidine deaminase, ZINC ION | | Authors: | Xie, K, Sowden, M.P, Dance, G.S.C, Torelli, A.T, Smith, H.C, Wedekind, J.E. | | Deposit date: | 2003-10-13 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1RAI
 
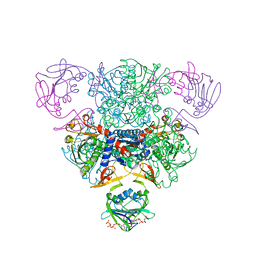 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
2DB4
 
 | | Crystal structure of rotor ring with DCCD of the V- ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DICYCLOHEXYLUREA, SODIUM ION, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7TT9
 
 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin C51S C71S Y34F variant | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Martinez, J.E, Liu, K, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2022-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N.
J.Biol.Chem., 301, 2025
|
|
2CFY
 
 | | Crystal structure of human thioredoxin reductase 1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN REDUCTASE 1 | | Authors: | Debreczeni, J.E, Johansson, C, Kavanagh, K, Savitsky, P, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Oppermann, U. | | Deposit date: | 2006-02-26 | | Release date: | 2006-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Thioredoxin Reductase 1
To be Published
|
|
2C7S
 
 | | Crystal structure of human protein tyrosine phosphatase kappa at 1.95A resolution | | Descriptor: | ACETATE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE KAPPA | | Authors: | Debreczeni, J.E, Ugochukwu, E, Eswaran, J, Barr, A, Das, S, Burgess, N, Gileadi, O, Longman, E, von Delft, F, Knapp, S, Sundstron, M, Arrowsmith, C, Weigelt, J, Edwards, A. | | Deposit date: | 2005-11-28 | | Release date: | 2007-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of human receptor protein tyrosine phosphatase kappa phosphatase domain 1.
Protein Sci., 15, 2006
|
|
2C60
 
 | | crystal structure of human mitogen-activated protein kinase kinase kinase 3 isoform 2 phox domain at 1.25 A resolution | | Descriptor: | CALCIUM ION, GLYCEROL, HUMAN MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 3 ISOFORM 2 | | Authors: | Debreczeni, J.E, Salah, E, Papagrigoriou, E, Burgess, N, von Delft, F, Gileadi, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Knapp, S. | | Deposit date: | 2005-11-04 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Human Mitogen-Activated Protein Kinase Kinase Kinase 3 Isoform 2 Fox Domain at 1.25 A Resolution
To be Published
|
|
2CLS
 
 | | The crystal structure of the human RND1 GTPase in the active GTP bound state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RHO-RELATED GTP-BINDING PROTEIN RHO6 | | Authors: | Pike, A.C.W, Yang, X, Colebrook, S, Gileadi, O, Sobott, F, Bray, J, Wen Hwa, L, Marsden, B, Zhao, Y, Schoch, G, Elkins, J, Debreczeni, J.E, Turnbull, A.P, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Crystal Structure of the Human Rnd1 Gtpase in the Active GTP Bound State
To be Published
|
|
2CA7
 
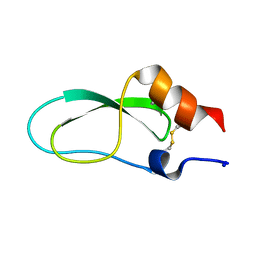 | | Conkunitzin-S1 Is The First Member Of A New Kunitz-Type Neurotoxin Family- Structural and Functional Characterization | | Descriptor: | CONKUNITZIN-S1 | | Authors: | Bayrhuber, M, Vijayan, V, Ferber, M, Graf, R, Korukottu, J, Imperial, J, Garrett, J.E, Olivera, B.M, Terlau, H, Zweckstetter, M, Becker, S. | | Deposit date: | 2005-12-19 | | Release date: | 2006-01-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Conkunitzin-S1 is the first member of a new Kunitz-type neurotoxin family. Structural and functional characterization.
J. Biol. Chem., 280, 2005
|
|
2C9W
 
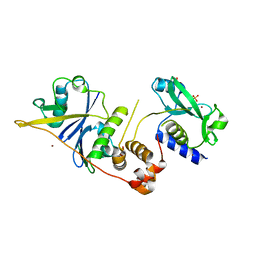 | | CRYSTAL STRUCTURE OF SOCS-2 IN COMPLEX WITH ELONGIN-B AND ELONGIN-C AT 1.9A RESOLUTION | | Descriptor: | NICKEL (II) ION, SULFATE ION, SUPPRESSOR OF CYTOKINE SIGNALING 2, ... | | Authors: | Debreczeni, J.E, Bullock, A, Amos, A, Savitsky, P, Barr, A, Burgess, N, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
2FGU
 
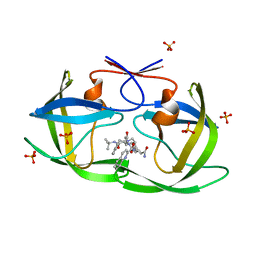 | | X-ray crystal structure of HIV-1 Protease T80S variant in complex with the inhibitor saquinavir used to explore the role of invariant Thr80 in HIV-1 protease structure, function, and viral infectivity. | | Descriptor: | (2S)-2-amino-3-phenylpropane-1,1-diol, 2-METHYL-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, ASPARAGINE, ... | | Authors: | Foulkes, J.E, Prabu-Jeyabalan, M, Cooper, D, Schiffer, C.A. | | Deposit date: | 2005-12-22 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity.
J.Virol., 80, 2006
|
|
2CYD
 
 | | Crystal structure of Lithium bound rotor ring of the V-ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, LITHIUM ION, UNDECYL-MALTOSIDE, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2006-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Lithium bound rotor ring of the V-ATPase from Enterococcus hirae
To be Published
|
|
1GMJ
 
 | | The structure of bovine IF1, the regulatory subunit of mitochondrial F-ATPase | | Descriptor: | ATPASE INHIBITOR | | Authors: | Cabezon, E, Runswick, M.J, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2001-09-14 | | Release date: | 2002-01-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Bovine If(1), the Regulatory Subunit of Mitochondrial F-ATPase.
Embo J., 20, 2001
|
|
2FGV
 
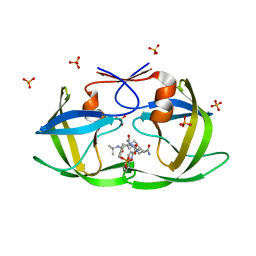 | | X-ray crystal structure of HIV-1 Protease T80N variant in complex with the inhibitor saquinavir used to explore the role of invariant Thr80 in HIV-1 protease structure, function, and viral infectivity. | | Descriptor: | (2S)-2-amino-3-phenylpropane-1,1-diol, 2-METHYL-DECAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, ASPARAGINE, ... | | Authors: | Foulkes, J.E, Prabu-Jeyabalan, M, Cooper, D, Schiffer, C.A. | | Deposit date: | 2005-12-22 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity.
J.Virol., 80, 2006
|
|
2FVL
 
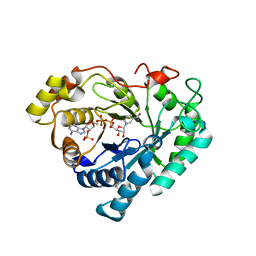 | | Crystal structure of human 3-alpha hydroxysteroid/dihydrodiol dehydrogenase (AKR1C4) complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1, member C4, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ugochukwu, E, Smee, C, Guo, K, Lukacik, P, Kavanagh, K, Debreczeni, J.E, von Delft, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human 3-alpha hydroxysteroid/dihydrodiol dehydrogenase (AKR1C4) complexed with NADP+
To be Published
|
|
2FSY
 
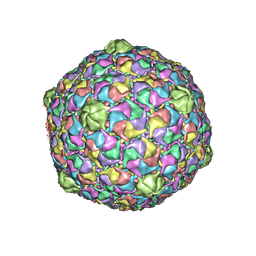 | | Bacteriophage HK97 Pepsin-treated Expansion Intermediate IV | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
2F6L
 
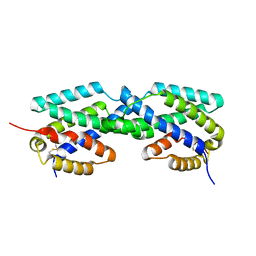 | | X-ray structure of Chorismate Mutase from Mycobacterium Tuberculosis | | Descriptor: | Chorismate mutase | | Authors: | Kim, S.K, Howard, A.J, Gilliland, G.L, Reddy, P.T, Ladner, J.E. | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of the secreted chorismate mutase (Rv1885c) from Mycobacterium tuberculosis H37Rv: an *AroQ enzyme not regulated by the aromatic amino acids.
J.Bacteriol., 188, 2006
|
|
2EZ6
 
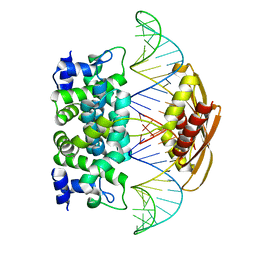 | | Crystal structure of Aquifex aeolicus RNase III (D44N) complexed with product of double-stranded RNA processing | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-11-10 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insight into the Mechanism of Double-Stranded RNA Processing by Ribonuclease III.
Cell(Cambridge,Mass.), 124, 2006
|
|
2FRP
 
 | | Bacteriophage HK97 Expansion Intermediate IV | | Descriptor: | Major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
2FT1
 
 | | Bacteriophage HK97 Head II | | Descriptor: | major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-23 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
2FS3
 
 | | Bacteriophage HK97 K169Y Head I | | Descriptor: | Major capsid protein | | Authors: | Gan, L, Speir, J.A, Conway, J.F, Lander, G, Cheng, N, Firek, B.A, Hendrix, R.W, Duda, R.L, Liljas, L, Johnson, J.E. | | Deposit date: | 2006-01-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Capsid Conformational Sampling in HK97 Maturation Visualized by X-Ray Crystallography and Cryo-EM.
Structure, 14, 2006
|
|
2D2V
 
 | |
1J9K
 
 | | CRYSTAL STRUCTURE OF SURE PROTEIN FROM T.MARITIMA IN COMPLEX WITH TUNGSTATE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, STATIONARY PHASE SURVIVAL PROTEIN, ... | | Authors: | Suh, S.W, Lee, J.Y, Kwak, J.E, Moon, J. | | Deposit date: | 2001-05-27 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family.
Nat.Struct.Biol., 8, 2001
|
|
