3BK5
 
 | |
3MW6
 
 | | Crystal structure of NMB1681 from Neisseria meningitidis MC58, a FinO-like RNA chaperone | | Descriptor: | GLYCEROL, uncharacterized protein NMB1681 | | Authors: | Tan, K, Zhou, M, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | N. meningitidis 1681 is a member of the FinO family of RNA chaperones.
Rna Biol., 7, 2010
|
|
3K0B
 
 | | Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, predicted N6-adenine-specific DNA methylase | | Authors: | Nocek, B, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a predicted N6-adenine-specific DNA methylase from Listeria monocytogenes str. 4b F2365
To be Published
|
|
3GN3
 
 | | Crystal structure of a putative protein-disulfide isomerase from Pseudomonas syringae to 2.5A resolution. | | Descriptor: | GLYCEROL, SULFATE ION, putative protein-disulfide isomerase | | Authors: | Stein, A.J, Chhor, G, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-16 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative protein-disulfide isomerase from Pseudomonas syringae to 2.5A resolution.
To be Published
|
|
1XWM
 
 | |
3NE8
 
 | | The crystal structure of a domain from N-acetylmuramoyl-l-alanine amidase of Bartonella henselae str. Houston-1 | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Rakowski, E, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division.
Mol.Microbiol., 85, 2012
|
|
3NEU
 
 | |
3N55
 
 | | SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, Peptidase, ... | | Authors: | Osipiuk, J, Mulligan, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-24 | | Release date: | 2010-06-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3BYW
 
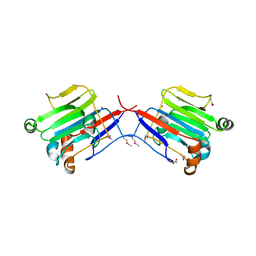 | | Crystal structure of an extracellular domain of arabinofuranosyltransferase from Corynebacterium diphtheriae | | Descriptor: | ACETATE ION, Putative arabinofuranosyltransferase, ZINC ION | | Authors: | Tan, K, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of an extracellular domain of arabinofuranosyltransferase from Corynebacterium diphtheriae.
To be Published
|
|
3NAT
 
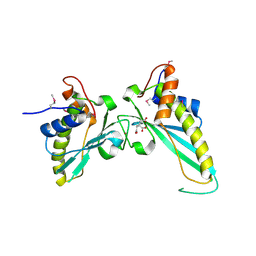 | | Crystal Structure of Conserved Protein of Unknown Function EF_1977 from Enterococcus faecalis | | Descriptor: | CITRIC ACID, Uncharacterized protein, ZINC ION | | Authors: | Kim, Y, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.925 Å) | | Cite: | Crystal Structure of Conserved Protein of Unknown Function EF_1977 from Enterococcus faecalis
To be Published
|
|
3C5O
 
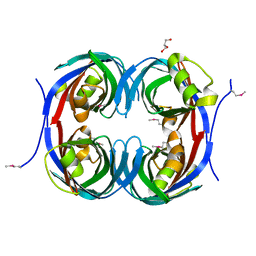 | | Crystal structure of the conserved protein of unknown function RPA1785 from Rhodopseudomonas palustris | | Descriptor: | GLYCEROL, UPF0311 protein RPA1785 | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-01 | | Release date: | 2008-02-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Conserved Protein of Unknown Function RPA1785 from Rhodopseudomonas palustris.
To be Published
|
|
4RW0
 
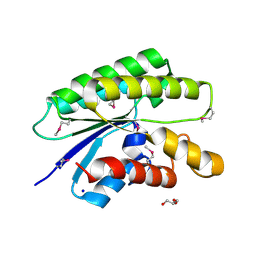 | | Crystal structure of a member of the lipolytic protein G-D-S-L family from Veillonella parvula DSM 2008 | | Descriptor: | GLYCEROL, Lipolytic protein G-D-S-L family, SODIUM ION | | Authors: | Nocek, B, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-30 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a member of the lipolytic protein G-D-S-L family from Veillonella parvula DSM 2008
To be Published
|
|
3BRC
 
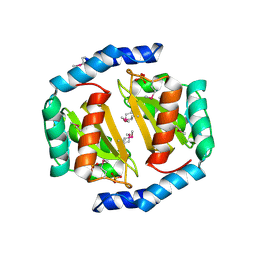 | | Crystal structure of a conserved protein of unknown function from Methanobacterium thermoautotrophicum | | Descriptor: | Conserved protein of unknown function, PHOSPHATE ION | | Authors: | Zhang, R, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a conserved protein of unknown function from Methanobacterium thermoautotrophicum.
To be Published
|
|
4PZJ
 
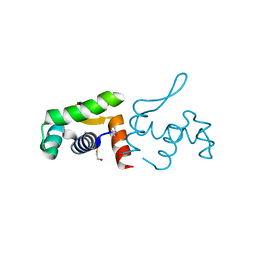 | | 1.60 Angstrom resolution crystal structure of a transcriptional regulator of the LysR family from Eggerthella lenta DSM 2243 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-31 | | Release date: | 2014-04-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of a transcriptional regulator of the LysR family from Eggerthella lenta DSM 2243
To be Published
|
|
3MVK
 
 | | The Crystal Structure of FucU from Bifidobacterium longum to 1.65A | | Descriptor: | GLYCEROL, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Stein, A.J, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of FucU from Bifidobacterium longum to 1.65A
To be Published
|
|
3C0U
 
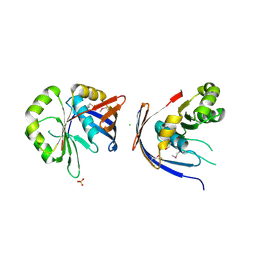 | | Crystal structure of E.coli yaeQ protein | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein yaeQ | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of E.coli yaeQ protein.
To be Published
|
|
3C6V
 
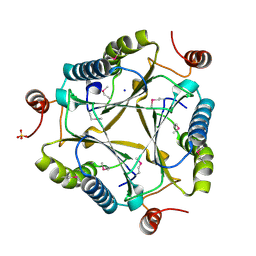 | | Crystal structure of AU4130/APC7354, a probable enzyme from the thermophilic fungus Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, Probable tautomerase/dehalogenase AU4130, SODIUM ION, ... | | Authors: | Singer, A.U, Binkowski, T.A, Skarina, T, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-05 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of AU4130/APC7354, a probable enzyme from the thermophilic fungus Aspergillus fumigatus.
To be Published
|
|
3NKH
 
 | | Crystal Structure of Integrase from MRSA strain Staphylococcus aureus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-19 | | Release date: | 2010-08-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal Structure of Integrase from MRSA strain Staphylococcus aureus
To be Published, 2010
|
|
1Y0U
 
 | | Crystal Structure of the putative arsenical resistance operon repressor from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, arsenical resistance operon repressor, putative | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-16 | | Release date: | 2004-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the putative arsenical resistance operon repressor from Archaeoglobus fulgidus
To be Published
|
|
3CEX
 
 | | Crystal structure of the conserved protein of locus EF_3021 from Enterococcus faecalis | | Descriptor: | ACETIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Cuff, M.E, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the conserved protein of locus EF_3021 from Enterococcus faecalis.
TO BE PUBLISHED
|
|
3K9U
 
 | | Crystal structure of paia acetyltransferase (ta0374) from thermoplasma acidophilum | | Descriptor: | ACETYL COENZYME *A, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
1Y89
 
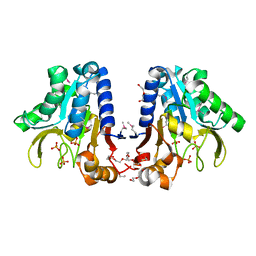 | | Crystal Structure of devB protein | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DI(HYDROXYETHYL)ETHER, NONAETHYLENE GLYCOL, ... | | Authors: | Lazarski, K, Cymborowski, M, Chruszcz, M, Zheng, H, Zhang, R, Lezondra, L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of devB protein
To be Published
|
|
4H0C
 
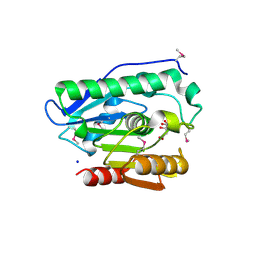 | | Crystal structure of phospholipase/Carboxylesterase from Dyadobacter fermentans DSM 18053 | | Descriptor: | CITRIC ACID, GLYCEROL, Phospholipase/Carboxylesterase, ... | | Authors: | Chang, C, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-07 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of phospholipase/Carboxylesterase from Dyadobacter fermentans DSM 18053
To be Published
|
|
3CZP
 
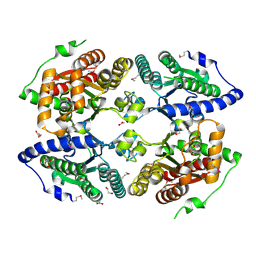 | | Crystal structure of putative polyphosphate kinase 2 from Pseudomonas aeruginosa PA01 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Nocek, B, Evdokimova, E, Osipiuk, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4KVH
 
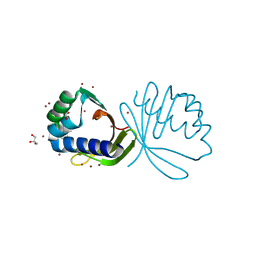 | | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747 | | Descriptor: | BROMIDE ION, CACODYLATE ION, FORMIC ACID, ... | | Authors: | Chang, C, Holowicki, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-22 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747
To be Published
|
|
