6YM3
 
 | | Crystal structure of Compound 1 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-ethoxyphenyl)phenyl]-5,8-dihydro-1,7-naphthyridin-4-yl]amino]propanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YM4
 
 | | Crystal structure of BAY-297 with PIP4K2A | | Descriptor: | (2~{R})-2-[[2-[4-(3-chloranyl-2-fluoranyl-phenyl)phenyl]-3-cyano-1,7-naphthyridin-4-yl]amino]butanamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
8ZSW
 
 | | Crystal Structure of Human DDB1, a Component of the E3 Ubiquitin Ligase Complex | | Descriptor: | ACETATE ION, DNA damage-binding protein 1 | | Authors: | Lee, J, Gil, Y, Jeong, Y.R, Jo, I. | | Deposit date: | 2024-06-05 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Improved-Resolution Crystal Structure of Human DDB1, a Component of the E3 Ubiquitin Ligase Complex
To Be Published
|
|
8ZDP
 
 | |
8ZDH
 
 | | Cryo-EM structure of Mycobacteriophage Douge genome-packed capsid (gp8 and gp113) | | Descriptor: | Capsid Cement Protein (gp113), Major Capsid Protein (gp8) | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
8ZDN
 
 | |
8ZDQ
 
 | | Cryo-EM structure of Mycobacteriophage Douge complete baseplate (gp13, gp17, gp23, gp16, gp18 and gp20) | | Descriptor: | Baseplate Hub Protein (gp18), Baseplate Upper Protein (gp23), Central Fiber Protein (gp20), ... | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
8ZDJ
 
 | | Cryo-EM structure of Mycobacteriophage Douge genome-packed connector (gp5, gp9, gp10, gp12 and gp13) | | Descriptor: | Adaptor Protein (gp9), Portal Protein (gp5), Stopper Protein (gp10), ... | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
8ZEA
 
 | |
8ZDI
 
 | |
8ZDO
 
 | | Cryo-EM structure of Mycobacteriophage Douge baseplate (gp13, gp17, gp23, gp16, gp18 and gp20) | | Descriptor: | Baseplate hub protein (gp18), Baseplate upper protein (gp23), Central fiber protein (gp20), ... | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
8ZYR
 
 | | Cryo-EM structure of BTN2A1-BTN3A1-BTN3A3 | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 2 member A1, Butyrophilin subfamily 3 member A1, ... | | Authors: | Gao, W, Zheng, J, Zhu, Y, Huang, Z. | | Deposit date: | 2024-06-18 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Cryo-EM structure of BTN2A1-BTN3A1-BTN3A3
To Be Published
|
|
8ZDL
 
 | | Cryo-EM structure of Mycobacteriophage Douge genome-free connector (gp5, gp9, gp10, gp12 and gp13) | | Descriptor: | Adaptor Protein (gp9), Protal Protein (gp5), Stopper Protein (gp10), ... | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
8ZWB
 
 | |
8ZYX
 
 | |
4UQJ
 
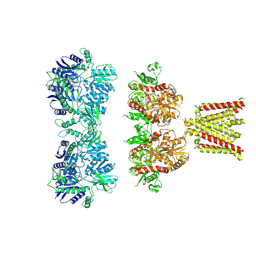 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
9B7F
 
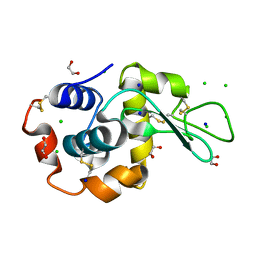 | | S_SAD structure of HEWL using lossless default compression | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Jakoncic, J, Bernstein, H.J, Soares, A.S, Horvat, K. | | Deposit date: | 2024-03-27 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Investigation of fast and efficient lossless compression algorithms for macromolecular crystallography experiments.
J.Synchrotron Radiat., 31, 2024
|
|
9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
9B7E
 
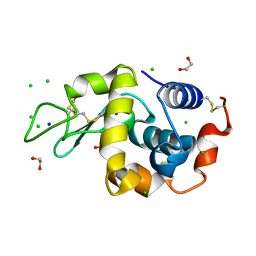 | | S_SAD structure of HEWL using lossy compression data with a compression ratio of 422 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Jakoncic, J, Bernstein, H.J, Soares, A.S, Horvat, K. | | Deposit date: | 2024-03-27 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Investigation of fast and efficient lossless compression algorithms for macromolecular crystallography experiments.
J.Synchrotron Radiat., 31, 2024
|
|
6Y4H
 
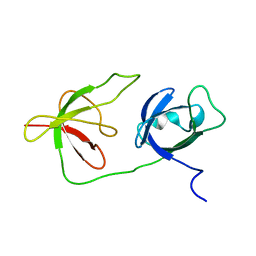 | |
6YU6
 
 | | Crystal structure of MhsT in complex with L-leucine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, LEUCINE, SODIUM ION, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
4UOM
 
 | | Electron Cryo-microscopy of Venezuelan Equine Encephalitis Virus TC- 83 in complex with neutralizing antibody Fab F5 | | Descriptor: | FAB FRAGMENT HEAVY CHAIN, FAB FRAGMENT LIGHT CHAIN | | Authors: | Porta, J, Jose, J, Roehrig, J.T, Blair, C.D, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2014-06-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Locking and Blocking the Viral Landscape of an Alphavirus with Neutralizing Antibodies.
J.Virol., 88, 2014
|
|
4UP4
 
 | | Structure of the recombinant lectin PVL from Psathyrella velutina in complex with GlcNAcb-D-1,3Galactoside | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Audfray, A, Beljoudi, M, Hurbin, A, Varrot, A, Breiman, A, Busser, B, Lependu, J, Coll, J.L, Imberty, A. | | Deposit date: | 2014-06-12 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Recombinant Fungal Lectin for Labeling Truncated Glycans on Human Cancer Cells.
Plos One, 10, 2015
|
|
4UPD
 
 | | Open conformation of O. piceae sterol esterase mutant I544W | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, STEROL ESTERASE, TRIETHYLENE GLYCOL, ... | | Authors: | Gutierrez-Fernandez, J, Vaquero, M.E, Prieto, A, Barriuso, J, Gonzalez, M.J, Hermoso, J.A. | | Deposit date: | 2014-06-16 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Ophiostoma Piceae Sterol Esterase: Structural Insights Into Activation Mechanism and Product Release.
J.Struct.Biol., 187, 2014
|
|
6YU4
 
 | | Crystal structure of MhsT in complex with L-4F-phenylalanine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4-FLUORO-L-PHENYLALANINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
