1PJG
 
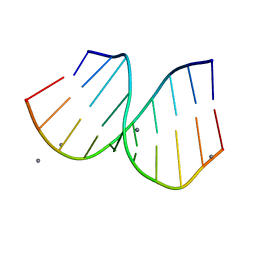 | | RNA/DNA Hybrid Decamer of CAAAGAAAAG/CTTTTCTTTG | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', CALCIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H.
J.Mol.Biol., 334, 2003
|
|
1I6Z
 
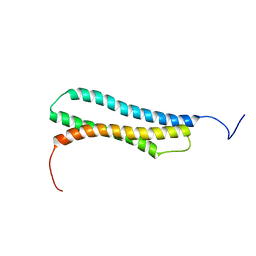 | | BAG DOMAIN OF BAG1 COCHAPERONE | | Descriptor: | BAG-FAMILY MOLECULAR CHAPERONE REGULATOR-1 | | Authors: | Briknarova, K, Takayama, S, Brive, L, Havert, M.L, Knee, D.A, Velasco, J, Homma, S, Cabezas, E, Stuart, J, Hoyt, D.W, Satterthwait, A.C, Llinas, M, Reed, J.C, Ely, K.R. | | Deposit date: | 2001-03-06 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein.
Nat.Struct.Biol., 8, 2001
|
|
1PJI
 
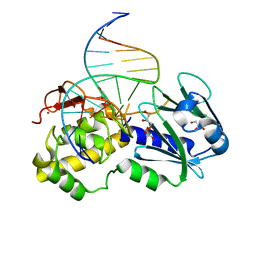 | | Crystal structure of wild type Lactococcus lactis FPG complexed to a 1,3 propanediol containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Pereira, K, Serre, L, Zelwer, C, Castaing, B. | | Deposit date: | 2003-06-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
1PJ4
 
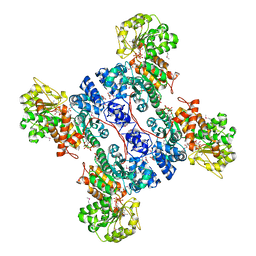 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, ATP, Mn++, and allosteric activator fumarate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MALATE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-31 | | Release date: | 2003-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
5UFX
 
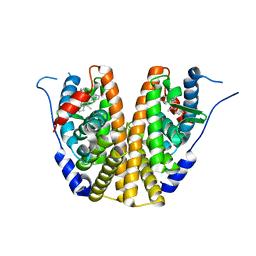 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with OP1074 | | Descriptor: | (2S)-3-(4-hydroxyphenyl)-4-methyl-2-(4-{2-[(3R)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-2H-1-benzopyran-7-ol, Estrogen receptor | | Authors: | Fanning, S.W, Hodges-Gallagher, L, Myles, D.C, Sun, R, Fowler, C.E, Green, B.D, Harmon, C.L, Greene, G.L, Kushner, P.J. | | Deposit date: | 2017-01-06 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5503 Å) | | Cite: | Specific stereochemistry of OP-1074 disrupts estrogen receptor alpha helix 12 and confers pure antiestrogenic activity.
Nat Commun, 9, 2018
|
|
1IB8
 
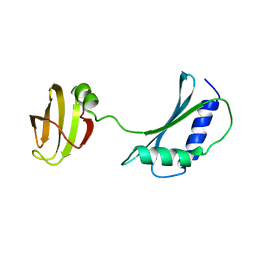 | | SOLUTION STRUCTURE AND FUNCTION OF A CONSERVED PROTEIN SP14.3 ENCODED BY AN ESSENTIAL STREPTOCOCCUS PNEUMONIAE GENE | | Descriptor: | CONSERVED PROTEIN SP14.3 | | Authors: | Yu, L, Gunasekera, A.H, Mack, J, Olejniczak, E.T, Chovan, L.E, Ruan, X, Towne, D.L, Lerner, C.G, Fesik, S.W. | | Deposit date: | 2001-03-27 | | Release date: | 2002-03-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE AND FUNCTION OF A CONSERVED PROTEIN SP14.3 ENCODED BY AN ESSENTIAL STREPTOCOCCUS PNEUMONIAE GENE
J.Mol.Biol., 311, 2001
|
|
5TV7
 
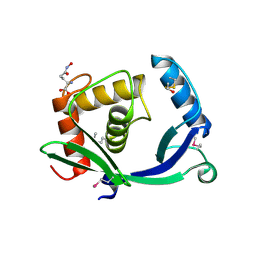 | | 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate. | | Descriptor: | GLUTAMINE HYDROXAMATE, Putative peptidoglycan-binding/hydrolysing protein | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Winsor, J, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate.
To Be Published
|
|
1I70
 
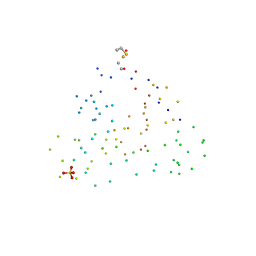 | | CRYSTAL STRUCTURE OF RNASE SA Y86F MUTANT | | Descriptor: | GUANYL-SPECIFIC RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Urbanikova, L. | | Deposit date: | 2001-03-07 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tyrosine hydrogen bonds make a large contribution to protein stability.
J.Mol.Biol., 312, 2001
|
|
1PJO
 
 | | Crystal Structure of an RNA/DNA hybrid of HIV-1 PPT | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', MAGNESIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An Unusual Sugar Conformation in the Structure of an RNA/DNA
Decamer of the Polypurine Tract May Affect Recognition by RNase H
J.Mol.Biol., 334, 2003
|
|
4NG8
 
 | | Dialyzed HEW lysozyme batch crystallized in 1.9 M CsCl and collected at 100 K. | | Descriptor: | CESIUM ION, CHLORIDE ION, Lysozyme C | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1PJ2
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, cofactor NADH, Mn++, and allosteric activator fumarate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
1PJ1
 
 | | RIBONUCLEOTIDE REDUCTASE R2-D84E/W48F SOAKED WITH FERROUS IONS AT PH 5 | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
1PM9
 
 | | CRYSTAL STRUCTURE OF HUMAN MNSOD H30N, Y166F MUTANT | | Descriptor: | MANGANESE (III) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Fan, L, Tainer, J.A. | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Amino acid substitution at the dimeric interface of human manganese superoxide dismutase
J.Biol.Chem., 279, 2004
|
|
1PJF
 
 | | Solid State NMR structure of the Pf1 Major Coat Protein in Magnetically Aligned Bacteriophage | | Descriptor: | COAT PROTEIN B | | Authors: | Thiriot, D.S, Nevzorov, A.A, Zagyanskiy, L, Wu, C.H, Opella, S.J. | | Deposit date: | 2003-06-02 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the coat protein in Pf1 bacteriophage determined by solid-state NMR spectroscopy.
J.Mol.Biol., 341, 2004
|
|
5U47
 
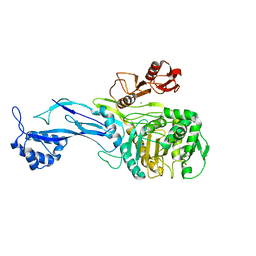 | | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus | | Descriptor: | ACETATE ION, CHLORIDE ION, Penicillin binding protein 2X | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-03 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus.
To Be Published
|
|
7QEN
 
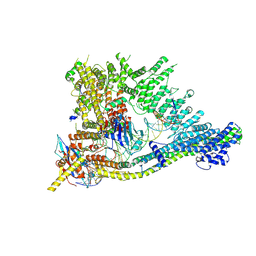 | | S.c. Condensin core in DNA- and ATP-bound state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Condensin complex subunit 1, Condensin complex subunit 2, ... | | Authors: | Lecomte, L, Hassler, M, Haering, C, Eustermann, S. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | A hold-and-feed mechanism drives directional DNA loop extrusion by condensin.
Science, 376, 2022
|
|
7QQK
 
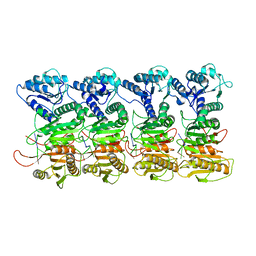 | | TIR-SAVED effector bound to cA3 | | Descriptor: | RNA (5'-R(P*AP*AP*A)-3'), TIR_SAVED fusion protein | | Authors: | Spagnolo, L, White, M.F, Hogrel, G, Guild, A. | | Deposit date: | 2022-01-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cyclic nucleotide-induced helical structure activates a TIR immune effector.
Nature, 608, 2022
|
|
1PKZ
 
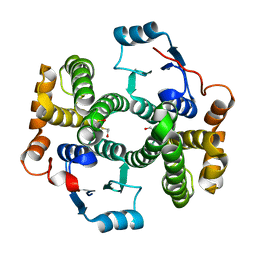 | | Crystal structure of human glutathione transferase (GST) A1-1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7QFW
 
 | | S.c. Condensin peripheral Ycg1 subcomplex bound to DNA | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, Synthetic DNA ligand, ... | | Authors: | Lecomte, L, Hassler, M, Haering, C, Eustermann, S. | | Deposit date: | 2021-12-06 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | A hold-and-feed mechanism drives directional DNA loop extrusion by condensin.
Science, 376, 2022
|
|
1HIY
 
 | | Binding of nucleotides to NDP kinase | | Descriptor: | 3'-DEOXY 3'-AMINO ADENOSINE-5'-DIPHOSPHATE, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Cervoni, L, Lascu, I, Xu, Y, Gonin, P, Morr, M, Merouani, M, Janin, J, Giartoso, A. | | Deposit date: | 2001-01-05 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of Nucleotides to Nucleoside Diphosphate Kinase: A Calorimetric Study.
Biochemistry, 40, 2001
|
|
1PQW
 
 | |
5UG4
 
 | | Structure of spermidine N-acetyltransferase SpeG from Vibrio cholerae | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-06 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of spermidine N-acetyltransferase SpeG from Vibrio cholerae
To Be Published
|
|
1PJ3
 
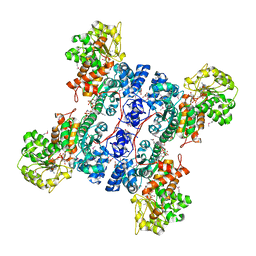 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate pyruvate, cofactor NAD+, Mn++, and allosteric activator fumarate. | | Descriptor: | FUMARIC ACID, MANGANESE (II) ION, NAD-dependent malic enzyme, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
5UHS
 
 | | Structure of a SemiSWEET D57A mutant | | Descriptor: | MONOVACCENIN, Sugar transporter SemiSWEET, beta-D-glucopyranose | | Authors: | Fastman, N.M, Feng, L. | | Deposit date: | 2017-01-11 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of Substrate Translocation in an Alternating Access Transporter.
Cell, 169, 2017
|
|
1PL4
 
 | | Crystal Structure of human MnSOD Y166F mutant | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Fan, L, Tainer, J.A. | | Deposit date: | 2003-06-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Amino acid substitution at the dimeric interface of human manganese superoxide dismutase
J.Biol.Chem., 279, 2004
|
|
