4GHI
 
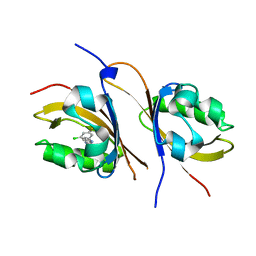 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains in complex with a benzoxadiazole antagonist | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(3-chloro-5-fluorophenyl)-4-nitro-2,1,3-benzoxadiazol-5-amine | | Authors: | Scheuermann, T.H, Key, J, Tambar, U.K, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2012-08-07 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Allosteric inhibition of hypoxia inducible factor-2 with small molecules.
Nat.Chem.Biol., 9, 2013
|
|
4J52
 
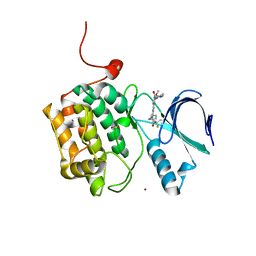 | | Crystal structure of PLK1 in complex with a pyrimidodiazepinone inhibitor | | Descriptor: | 4-{[(7R)-9-cyclopentyl-7-ethenyl-7-fluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl]amino}-3-methoxy-N-(4-methylpiperazin-1-yl)benzamide, ACETATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Hosfield, D.J, Skene, R.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of TAK-960: An orally available small molecule inhibitor of polo-like kinase 1 (PLK1).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4J53
 
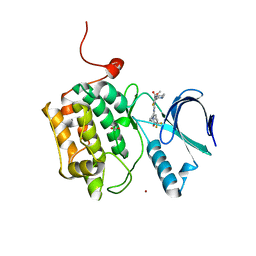 | | Crystal structure of PLK1 in complex with TAK-960 | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-2-fluoro-5-methoxy-N-(1-methylpiperidin-4-yl)benzamide, ACETATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Hosfield, D.J, Skene, R.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of TAK-960: An orally available small molecule inhibitor of polo-like kinase 1 (PLK1).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6LEI
 
 | |
6LHH
 
 | | Crystal structure of chicken 8mer-BF2*1501 | | Descriptor: | ARG-ARG-ARG-GLU-GLN-THR-ASP-TYR, Beta-2-microglobulin, MHC class I | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
6LHF
 
 | | Crystal structure of chicken cCD8aa/pBF2*15:01 | | Descriptor: | Beta-2-microglobulin, CD8 alpha chain, MHC class I, ... | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
6U0S
 
 | | Crystal structure of the flavin-dependent monooxygenase PieE in complex with FAD and substrate | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-6-methoxy-3-methylpyridin-4-ol, CHLORIDE ION, ... | | Authors: | Shi, R, Manenda, M. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6U0P
 
 | | Crystal structure of PieE, the flavin-dependent monooxygenase involved in the biosynthesis of piericidin A1 | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Shi, R, Manenda, M, Picard, M.-E. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
6LED
 
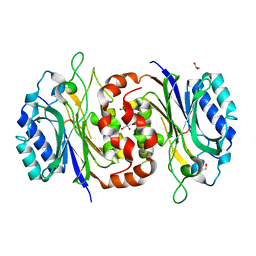 | |
6LHG
 
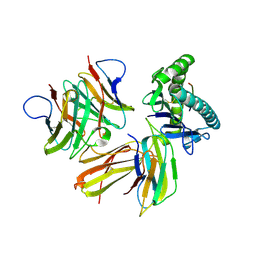 | | Crystal structure of chicken cCD8aa/pBF2*04:01 | | Descriptor: | Beta-2-microglobulin, CD8 alpha chain, IE8 peptide, ... | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
6LE2
 
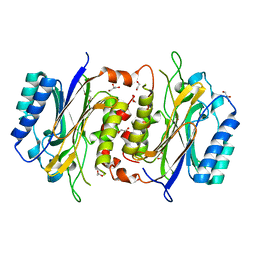 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-carbamoyl-D-amino-acid hydrolase | | Authors: | Ni, Y, Liu, Y.F, Xu, G.C, Dai, W. | | Deposit date: | 2019-11-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
6ILG
 
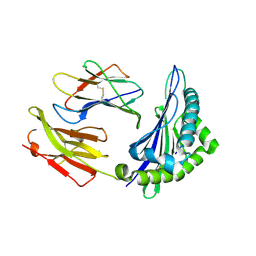 | |
6ILF
 
 | |
6VPX
 
 | |
6N7R
 
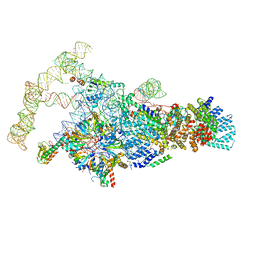 | | Saccharomyces cerevisiae spliceosomal E complex (ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA, Pre-mRNA-processing factor 39, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-28 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A unified mechanism for intron and exon definition and back-splicing.
Nature, 573, 2019
|
|
6NAE
 
 | |
6NCU
 
 | | Interleukin-37 residues 53-206- dimer | | Descriptor: | Interleukin-37 | | Authors: | Eisenmesser, E.Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-03-13 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Interleukin-37 monomer is the active form for reducing innate immunity.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6N7P
 
 | | S. cerevisiae spliceosomal E complex (UBC4) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Nuclear cap-binding protein complex subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-27 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A unified mechanism for intron and exon definition and back-splicing.
Nature, 573, 2019
|
|
6NX1
 
 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN BOUND TETHERED WITH SRC CO-ACTIVATOR PEPTIDE AND COMPOUND-3 AKA 1,1,1,3,3,3-HEXAFLUORO-2-{4-[1-(4- LUOROBENZENESULFONYL)CYCLOPENTYL]PHENYL}PROPAN-2-OL | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-(4-{1-[(4-fluorophenyl)sulfonyl]cyclopentyl}phenyl)propan-2-ol, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 fusion | | Authors: | Khan, J.A. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-based Discovery of Phenyl (3-Phenylpyrrolidin-3-yl)sulfones as Selective, Orally Active ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 10, 2019
|
|
6ILE
 
 | | CRYSTAL STRUCTURE OF A MUTANT PTAL-N*01:01 FOR 2.9 ANGSTROM, 52M 53D 54L DELETED | | Descriptor: | Beta-2-microglobulin, HEV-1, MHC class I antigen | | Authors: | Qu, Z.H, Zhang, N.Z, Xia, C. | | Deposit date: | 2018-10-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Peptidome of the Bat MHC Class I Molecule Reveal a Novel Mechanism Leading to High-Affinity Peptide Binding.
J Immunol., 202, 2019
|
|
6ILC
 
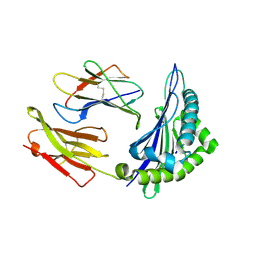 | |
6QVC
 
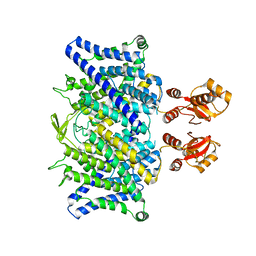 | |
5ZMA
 
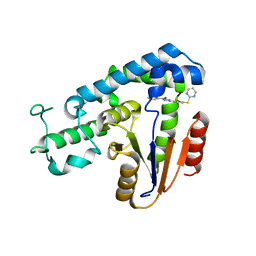 | | Structural basis for an allosteric Eya2 phosphatase inhibitor | | Descriptor: | 3-fluoro-N'-[(E)-{5-[(pyrimidin-2-yl)sulfanyl]furan-2-yl}methylidene]benzohydrazide, Eyes absent homolog 2 | | Authors: | Anantharajan, J, Jansson, A.E, Kang, C. | | Deposit date: | 2018-04-02 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.175 Å) | | Cite: | Structural and Functional Analyses of an Allosteric EYA2 Phosphatase Inhibitor That Has On-Target Effects in Human Lung Cancer Cells.
Mol.Cancer Ther., 18, 2019
|
|
4KC3
 
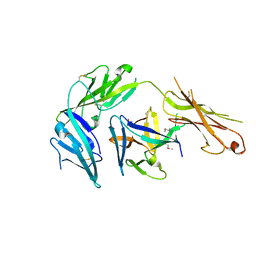 | | Cytokine/receptor binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor-like 1, Interleukin-33 | | Authors: | Liu, X, Wang, X.Q. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2702 Å) | | Cite: | Structural insights into the interaction of IL-33 with its receptors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LWG
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ4 | | Descriptor: | 1-(5-chloro-2,4-dihydroxyphenyl)-2-(4-methoxyphenyl)ethanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Shi, F, Xiong, B, He, J. | | Deposit date: | 2013-07-27 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
