2F0X
 
 | | Crystal structure and function of human thioesterase superfamily member 2(THEM2) | | Descriptor: | SULFATE ION, Thioesterase superfamily member 2 | | Authors: | Cheng, Z, Song, F, Shan, X, Wang, Y, Wei, Z, Gong, W. | | Deposit date: | 2005-11-14 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human thioesterase superfamily member 2
Biochem.Biophys.Res.Commun., 349, 2006
|
|
3OEE
 
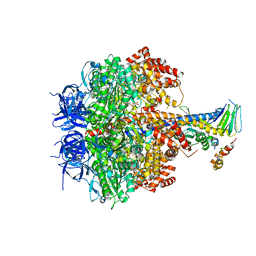 | | Structure of four mutant forms of yeast F1 ATPase: alpha-F405S | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of four mutant forms of yeast F1 ATPase: alpha-F405S
To be Published
|
|
3OFN
 
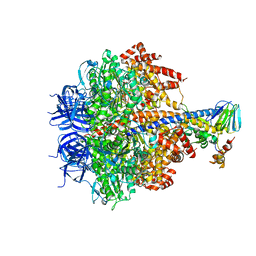 | | Structure of four mutant forms of yeast F1 ATPase: alpha-N67I | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
7CAH
 
 | | The interface of H014 Fab binds to SARS-CoV-2 S | | Descriptor: | Heavy chain of H014 Fab, Light chain of H014 Fab, Spike protein S1 | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Rao, Z, wang, Y, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAK
 
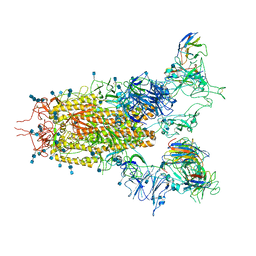 | | SARS-CoV-2 S trimer with three RBD in the open state and complexed with three H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
3OXZ
 
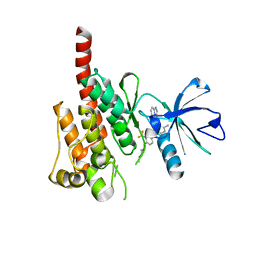 | | Crystal structure of ABL kinase domain bound with a DFG-out inhibitor AP24534 | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
6KZC
 
 | | crystal structure of TRKc in complex with 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-N-(3-((4- methylpiperazin-1-yl)methyl)-5- (trifluoromethyl)phenyl)benzamide | | Descriptor: | 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]benzamide, NT-3 growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
3OY3
 
 | | Crystal structure of ABL T315I mutant kinase domain bound with a DFG-out inhibitor AP24589 | | Descriptor: | 5-[(5-{[4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-3-(trifluoromethyl)phenyl]carbamoyl}-2-methylphenyl)ethynyl]-1-methyl-1H-imidazole-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Commodore, L, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
3OU3
 
 | | MDR769 HIV-1 protease complexed with PR/RT hepta-peptide | | Descriptor: | HIV-1 protease, PR/RT substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OUB
 
 | | MDR769 HIV-1 protease complexed with NC/p1 hepta-peptide | | Descriptor: | MDR HIV-1 protease, NC/p1 substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
2HVX
 
 | | Discovery of Potent, Orally Active, Nonpeptide Inhibitors of Human Mast Cell Chymase by Using Structure-Based Drug Design | | Descriptor: | Chymase, [(1S)-1-(5-CHLORO-1-BENZOTHIEN-3-YL)-2-(2-NAPHTHYLAMINO)-2-OXOETHYL]PHOSPHONIC ACID | | Authors: | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond, H.R, de Garavilla, L, Wang, Y, Minor, L.A, Wells, G.I, Di Cera, E, Cantwell, A.M, Savvides, S.N, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2006-07-31 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of potent, selective, orally active, nonpeptide inhibitors of human mast cell chymase.
J.Med.Chem., 50, 2007
|
|
4E6K
 
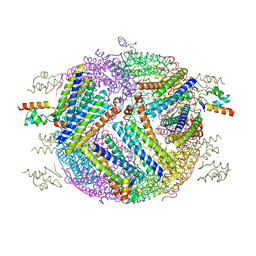 | | 2.0 A resolution structure of Pseudomonas aeruginosa bacterioferritin (BfrB) in complex with bacterioferritin associated ferredoxin (Bfd) | | Descriptor: | Bacterioferritin, FE2/S2 (INORGANIC) CLUSTER, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Wang, Y, Kumar, R, Ruvinsky, A, Vasker, I, Rivera, M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin.
J.Am.Chem.Soc., 134, 2012
|
|
4EG2
 
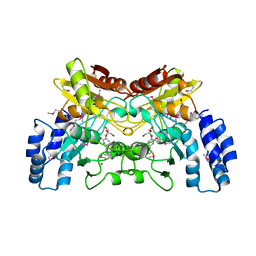 | | 2.2 Angstrom Crystal Structure of Cytidine deaminase from Vibrio cholerae in Complex with Zinc and Uridine | | Descriptor: | ACETATE ION, Cytidine deaminase, MAGNESIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Wang, Y, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Cytidine deaminase from Vibrio cholerae in Complex with Zinc and Uridine.
TO BE PUBLISHED
|
|
7V5K
 
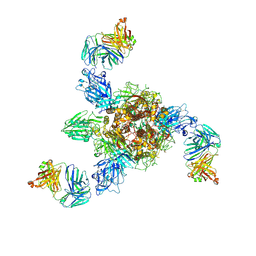 | | MERS S ectodomain trimer in complex with neutralizing antibody 0722 (state 1) | | Descriptor: | 0722 H, 0722 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 0722 (state 1)
to be published
|
|
7V5J
 
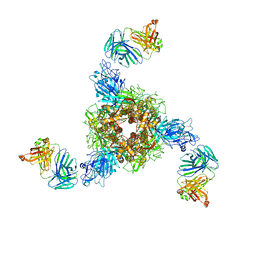 | | MERS S ectodomain trimer in complex with neutralizing antibody 0722(state 2) | | Descriptor: | 0722 H, 0722 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 0722(state 2)
to be published
|
|
7VIB
 
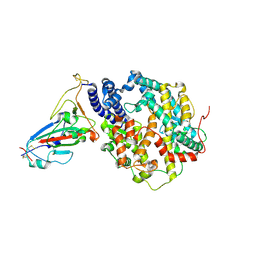 | | Crystal structure of human ACE2 and GX/P2V RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein, ZINC ION | | Authors: | Guo, Y, Cao, W, Jia, N, Wang, W, Yuan, S, Wang, Y. | | Deposit date: | 2021-09-26 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human ACE2 and GX/P2V RBD
To Be Published
|
|
6LWU
 
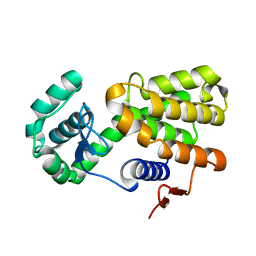 | | Complex of Gynuella sunshinyii GH46 chitosanase GsCsn46A E19A with chitopentaose | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, chitosanase | | Authors: | Qin, Z, Wang, Y. | | Deposit date: | 2020-02-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.419 Å) | | Cite: | Complex of Gynuella sunshinyii GH46 chitosanase GsCsn46A E19A with chitopentaose
To Be Published
|
|
8GRX
 
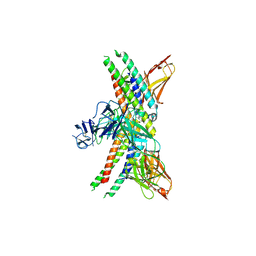 | | APOE4 receptor in complex with APOE4 NTD | | Descriptor: | Apolipoprotein E, Leukocyte immunoglobulin-like receptor subfamily A member 6 | | Authors: | Zhou, J, Wang, Y, Huang, G, Shi, Y. | | Deposit date: | 2022-09-02 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | LilrB3 is a putative cell surface receptor of APOE4.
Cell Res., 33, 2023
|
|
4DZD
 
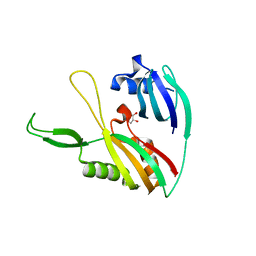 | |
6NOH
 
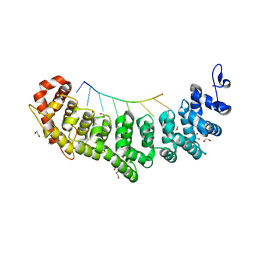 | | Crystal structure of FBF-2 repeat 5 mutant (C363S, R364Y, Q367S) in complex with 8-nt RNA | | Descriptor: | 1,2-ETHANEDIOL, Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*GP*UP*AP*AP*AP*UP*A)-3') | | Authors: | McCann, K.L, Wang, Y, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
3B96
 
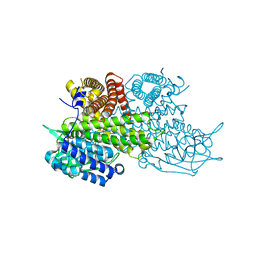 | | Structural Basis for Substrate Fatty-Acyl Chain Specificity: Crystal Structure of Human Very-Long-Chain Acyl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TETRADECANOYL-COA, Very long-chain specific acyl-CoA dehydrogenase | | Authors: | McAndrew, R.P, Wang, Y, Mohsen, A.W, He, M, Vockley, J, Kim, J.J. | | Deposit date: | 2007-11-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
6NOC
 
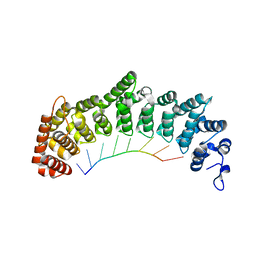 | | Crystal structure of FBF-2 repeat 5 mutant (C363A, R364Y) in complex with 8-nt RNA | | Descriptor: | Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*GP*UP*AP*AP*AP*UP*A)-3') | | Authors: | McCann, K, Wang, Y, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
6NOF
 
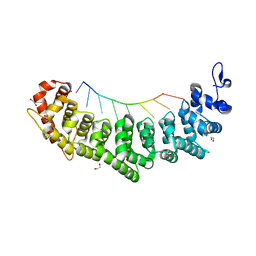 | | Crystal structure of FBF-2 repeat 5 mutant (C363A, R364Y, Q367S) in complex with 8-nt RNA | | Descriptor: | 1,2-ETHANEDIOL, Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*GP*UP*AP*AP*AP*UP*A)-3') | | Authors: | McCann, K.L, Wang, Y, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
8GXD
 
 | | L-LEUCINE DEHYDROGENASE FROM EXIGUOBACTERIUM SIBIRICUM | | Descriptor: | CALCIUM ION, GLYCEROL, Glu/Leu/Phe/Val dehydrogenase | | Authors: | Mu, X, Nie, Y, Wu, T, Wang, Y, Zhang, N, Yin, D, Xu, Y. | | Deposit date: | 2022-09-19 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Reshaping Substrate-Binding Pocket of Leucine Dehydrogenase for Bidirectionally Accessing Structurally Diverse Substrates
Acs Catalysis, 13, 2023
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
