8K1O
 
 | | mycobacterial efflux pump, AMPPNP bound state | | Descriptor: | CARDIOLIPIN, MAGNESIUM ION, Multidrug efflux system ATP-binding protein Rv1218c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8K1M
 
 | | mycobacterial efflux pump, apo state | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CARDIOLIPIN, Multidrug efflux system ATP-binding protein Rv1218c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8K1N
 
 | | mycobacterial efflux pump, substrate-bound state | | Descriptor: | CARDIOLIPIN, Multidrug efflux system ATP-binding protein Rv1218c, Multidrug efflux system permease protein Rv1217c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7BWR
 
 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein, ... | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
5WYM
 
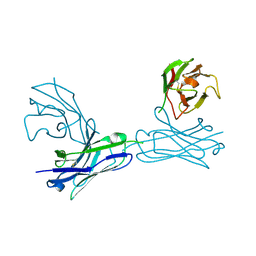 | | Crystal structure of an anti-connexin26 scFv | | Descriptor: | anti-connexin26 scFv,Ig heavy chain,Linker,anti-connexin26 scFv,Ig light chain | | Authors: | Li, S, Xu, L. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design and Characterization of a Human Monoclonal Antibody that Modulates Mutant Connexin 26 Hemichannels Implicated in Deafness and Skin Disorders
Front Mol Neurosci, 10, 2017
|
|
7VYW
 
 | |
7VZ2
 
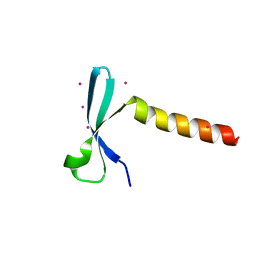 | | Crystal structure of chromodomain of Arabidopsis LHP1 | | Descriptor: | Chromo domain-containing protein LHP1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Min, J. | | Deposit date: | 2021-11-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the recognition of methylated histone H3 by the Arabidopsis LHP1 chromodomain.
J.Biol.Chem., 298, 2022
|
|
8HTV
 
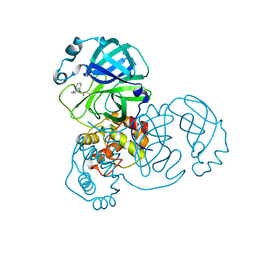 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3a | | Descriptor: | 1-(5,6-dihydrobenzo[b][1]benzazepin-11-yl)-2-sulfanyl-ethanone, 3C-like proteinase | | Authors: | Su, H.X, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Mechanism Study of SARS-CoV-2 3C-like Protease Inhibitors with a New Reactive Group.
J.Med.Chem., 66, 2023
|
|
7E0B
 
 | |
4HWH
 
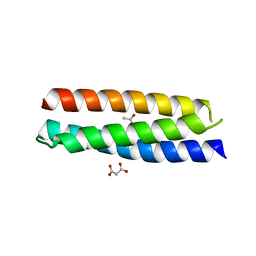 | | Crystal structure of ATBAG4 | | Descriptor: | ACETATE ION, BAG family molecular chaperone regulator 4, GLYCEROL, ... | | Authors: | Shen, Y, Fang, S. | | Deposit date: | 2012-11-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structural insight into plant programmed cell death mediated by BAG proteins in Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8K4F
 
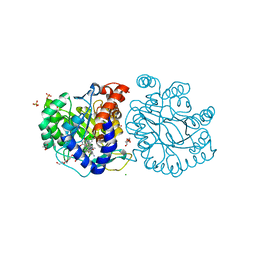 | | DHODH in complex with compound A0 | | Descriptor: | 5-cyclopropyl-2-[1-[(2-fluorophenyl)methyl]pyrazolo[3,4-b]pyridin-3-yl]pyrimidin-4-amine, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, ACETATE ION, ... | | Authors: | Jian, L, Sun, Q. | | Deposit date: | 2023-07-18 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery and Optimization of Novel h DHODH Inhibitors for the Treatment of Inflammatory Bowel Disease.
J.Med.Chem., 66, 2023
|
|
5ZX1
 
 | |
4HWI
 
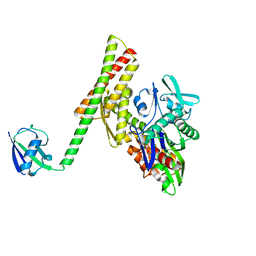 | | Crystal structure of ATBAG1 in complex with HSP70 | | Descriptor: | BAG family molecular chaperone regulator 1, Heat shock cognate 71 kDa protein | | Authors: | Shen, Y, Fang, S. | | Deposit date: | 2012-11-07 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structural insight into plant programmed cell death mediated by BAG proteins in Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HWD
 
 | | Crystal structure of ATBAG2 | | Descriptor: | BAG family molecular chaperone regulator 2 | | Authors: | Shen, Y, Fang, S. | | Deposit date: | 2012-11-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural insight into plant programmed cell death mediated by BAG proteins in Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6A73
 
 | | Complex structure of CSN2 with IP6 | | Descriptor: | COP9 signalosome complex subunit 2,Endolysin, INOSITOL HEXAKISPHOSPHATE, SULFATE ION | | Authors: | Liu, L, Li, D, Rao, F, Wang, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8ZBE
 
 | | cryo-EM structure of the octreotide-bound SSTR5-Gi complex | | Descriptor: | Beta-2 adrenergic receptor,Somatostatin receptor type 5,lgbit (fusion protein), Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Li, Y.G, Meng, X.Y, Yang, X.R, Ling, S.L, Shi, P, Tian, C.L, Yang, F. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into somatostatin receptor 5 bound with cyclic peptides.
Acta Pharmacol.Sin., 45, 2024
|
|
8ZCJ
 
 | | Cryo-EM structure of the pasireotide-bound SSTR5-Gi complex | | Descriptor: | 004-DTR-LYS-TYR-PHA-HYP, Beta-2 adrenergic receptor,Somatostatin receptor type 5,lgbit (fusion protein), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y.G, Meng, X.Y, Yang, X.R, Ling, S.L, Shi, P, Tian, C.L, Yang, F. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into somatostatin receptor 5 bound with cyclic peptides.
Acta Pharmacol.Sin., 45, 2024
|
|
4HWF
 
 | | Crystal structure of ATBAG3 | | Descriptor: | BAG family molecular chaperone regulator 3, SULFATE ION | | Authors: | Shen, Y, Fang, S. | | Deposit date: | 2012-11-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into plant programmed cell death mediated by BAG proteins in Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HWC
 
 | | Structure of ATBAG1 | | Descriptor: | BAG family molecular chaperone regulator 1, CHLORIDE ION | | Authors: | Shen, Y, Fang, S. | | Deposit date: | 2012-11-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural insight into plant programmed cell death mediated by BAG proteins in Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4LH9
 
 | |
7XEZ
 
 | |
7YGI
 
 | | Crystal structure of p53 DBD domain in complex with azurin | | Descriptor: | Azurin, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Jiang, W.X, Zuo, J.Q, Hu, J.J, Chen, X.Q, Ma, L.X, Liu, Z, Xing, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of bacterial effector protein azurin targeting tumor suppressor p53 and inhibiting its ubiquitination.
Commun Biol, 6, 2023
|
|
6DBK
 
 | | Tyk2 with compound 8 | | Descriptor: | 4-({4-[(1S,4S)-5-(cyanoacetyl)-2,5-diazabicyclo[2.2.1]heptan-2-yl]pyrimidin-2-yl}amino)-N-ethylbenzamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of (( S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841).
J. Med. Chem., 61, 2018
|
|
