8XW3
 
 | |
8XW1
 
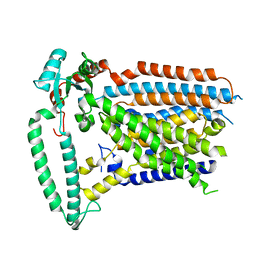 | | Cryo-EM structure of OSCA1.2-V335W-DDM state | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.49 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XVY
 
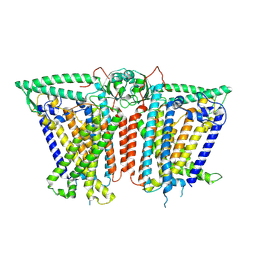 | |
8XW2
 
 | |
8XRY
 
 | |
8XS5
 
 | |
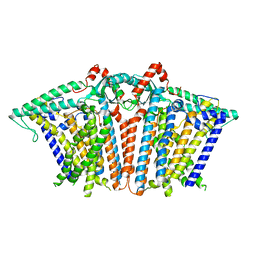
8XW0
 
 | | Cryo-EM structure of OSCA3.1-GDN state | | Descriptor: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XVZ
 
 | |
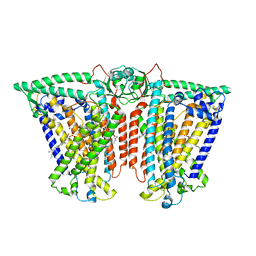
8XW4
 
 | | Cryo-EM structure of TMEM63B-Digitonin state | | Descriptor: | CSC1-like protein 2 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XVX
 
 | |
8XS0
 
 | |
7PB2
 
 | | Crystal structure of JDI TCR in complex with HLA-A*11:01 bound to KRAS G12D peptide (VVVGADGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS G12D peptide (VVVGADGVGK), MHC class I antigen, ... | | Authors: | Coles, C.H, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Therapeutic high affinity T cell receptor targeting a KRASG12D cancer neoantigen
Nat Commun, 13, 2022
|
|
7OW5
 
 | |
7OW6
 
 | |
7OW3
 
 | |
7OW4
 
 | | Crystal structure of HLA-A*11:01 in complex with KRAS G12D peptide (VVVGADGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS G12D peptide (VVVGADGVGK), MHC class I antigen, ... | | Authors: | Coles, C.H, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2021-06-16 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Therapeutic high affinity T cell receptor targeting a KRASG12D cancer neoantigen
Nat Commun, 13, 2022
|
|
3G36
 
 | | Crystal structure of the human DPY-30-like C-terminal domain | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Wang, X, Lou, Z, Bartlam, M, Rao, Z. | | Deposit date: | 2009-02-02 | | Release date: | 2009-06-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the C-terminal domain of human DPY-30-like protein: A component of the histone methyltransferase complex
J.Mol.Biol., 390, 2009
|
|
3DBE
 
 | |
3DBD
 
 | |
3DBC
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 257 | | Descriptor: | 3-[3-(3-methyl-6-{[(1S)-1-phenylethyl]amino}-1H-pyrazolo[4,3-c]pyridin-1-yl)phenyl]propanamide, Polo-like kinase 1 | | Authors: | Elling, R.A, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-31 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Design and synthesis of 2-amino-pyrazolopyridines as Polo-like kinase 1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DBF
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 562 | | Descriptor: | 4-({1-[3-(3-amino-3-oxopropyl)-5-chlorophenyl]-3-methyl-1H-pyrazolo[4,3-c]pyridin-6-yl}amino)-3-methoxy-N-(1-methylpipe ridin-4-yl)benzamide, Polo-like kinase | | Authors: | Elling, R.A, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-31 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 2-amino-pyrazolopyridines as Polo-like kinase 1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5NCY
 
 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-(3-methyl-5-oxidanylidene-6-phenyl-[1,3]thiazolo[3,2-a]pyridin-7-yl)ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ... | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
5NCZ
 
 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-[6-[3-[(dimethylamino)methyl]phenyl]-3-methyl-5-oxidanylidene-[1,3]thiazolo[3,2-a]pyridin-7-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
5GMZ
 
 | | Hepatitis B virus core protein Y132A mutant in complex with 4-methyl heteroaryldihydropyrimidine | | Descriptor: | (2S)-4,4-difluoro-1-[[(4S)-4-(4-fluorophenyl)-5-methoxycarbonyl-4-methyl-2-(1,3-thiazol-2-yl)-1H-pyrimidin-6-yl]methyl]pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors
J.Med.Chem., 59, 2016
|
|
