6Z87
 
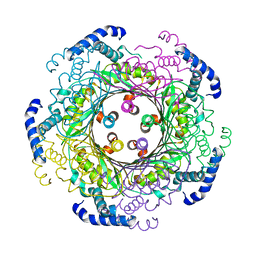 | | human GTP cyclohydrolase I | | Descriptor: | GTP cyclohydrolase 1, ZINC ION | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4UIE
 
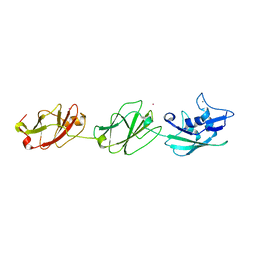 | | Crystal structure of the S-layer protein SbsC, domains 7, 8 and 9 | | Descriptor: | CALCIUM ION, OSMIUM ION, SURFACE LAYER PROTEIN | | Authors: | Pavkov-Keller, T, Dordic, A, Egelseer, E.M, Sleytr, U.B, Keller, W. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the S-Layer Protein Sbsc
To be Published
|
|
4UID
 
 | |
4UIC
 
 | |
3O20
 
 | | Electron transfer complexes:experimental mapping of the Redox-dependent Cytochrome C electrostatic surface | | Descriptor: | Cytochrome c, HEME C, NITRATE ION | | Authors: | De March, M, De Zorzi, R, Casini, A, Messori, L, Geremia, S, Demitri, N, Gabbiani, C, Guerri, A. | | Deposit date: | 2010-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nitrate as a probe of cytochrome c surface: crystallographic identification of crucial "hot spots" for protein-protein recognition.
J. Inorg. Biochem., 135, 2014
|
|
8AD3
 
 | | X-ray structure of NqrF(129-408)of Vibrio cholerae variant F406A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Na(+)-translocating NADH-quinone reductase subunit F | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ACW
 
 | | X-ray structure of Na+-NQR from Vibrio cholerae at 3.4 A resolution | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ACY
 
 | | X-ray structure of Na+-NQR from Vibrio cholerae at 3.5 A resolution | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AD4
 
 | | X-ray structure of NqrF(129-408)of Vibrio cholerae in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AD5
 
 | | X-ray structure of NqrF(129-408)of Vibrio cholerae variant F406A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5FL7
 
 | | Structure of the F1c10 complex from Yarrowia lipolytica ATP synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP SYNTHASE DELTA CHAIN, ... | | Authors: | Parey, K, Bublitz, M, Meier, T. | | Deposit date: | 2015-10-22 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a Complete ATP Synthase Dimer Reveals the Molecular Basis of Inner Mitochondrial Membrane Morphology.
Mol.Cell, 63, 2016
|
|
3O1Y
 
 | | Electron transfer complexes: Experimental mapping of the redox-dependent cytochrome c electrostatic surface | | Descriptor: | Cytochrome c, HEME C, NITRATE ION | | Authors: | De March, M, De Zorzi, R, Demitri, N, Gabbiani, C, Guerri, A, Casini, A, Messori, L, Geremia, S. | | Deposit date: | 2010-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nitrate as a probe of cytochrome c surface: crystallographic identification of crucial "hot spots" for protein-protein recognition.
J. Inorg. Biochem., 135, 2014
|
|
7ACC
 
 | |
7AL9
 
 | |
7ALA
 
 | | human GCH-GFRP inhibitory complex | | Descriptor: | 2-azanyl-8-[(4-fluorophenyl)methylsulfanyl]-1,7-dihydropurin-6-one, GTP cyclohydrolase 1, GTP cyclohydrolase 1 feedback regulatory protein, ... | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ALC
 
 | | human GCH-GFRP stimulatory complex | | Descriptor: | GTP cyclohydrolase 1, GTP cyclohydrolase 1 feedback regulatory protein, PHENYLALANINE, ... | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ALB
 
 | | human GCH-GFRP stimulatory complex 7-deaza-GTP bound | | Descriptor: | 7-deaza-GTP, GTP cyclohydrolase 1, GTP cyclohydrolase 1 feedback regulatory protein, ... | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NY5
 
 | | The apo structure of 3,4-dihydroxybenzoic acid decarboxylases from Enterobacter cloacae | | Descriptor: | 3,4-dihydroxybenzoate decarboxylase, GLYCEROL | | Authors: | Dordic, A, Gruber, K, Payer, S, Glueck, S, Pavkov-Keller, T, Marshall, S, Leys, D. | | Deposit date: | 2017-05-11 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6RKE
 
 | | Molybdenum storage protein - P212121, ADP, molybdate | | Descriptor: | (mu3-oxo)-tris(mu2-oxo)-nonakisoxo-trimolybdenum (VI), ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ermler, U, Bruenle, S. | | Deposit date: | 2019-04-30 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molybdate pumping into the molybdenum storage protein via an ATP-powered piercing mechanism.
Proc.Natl.Acad.Sci.USA, 2019
|
|
6RIS
 
 | | The Kb42S variant of the molybdenum storage protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ermler, U, Bruenle, S. | | Deposit date: | 2019-04-25 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molybdate pumping into the molybdenum storage protein via an ATP-powered piercing mechanism.
Proc.Natl.Acad.Sci.USA, 2019
|
|
6RJ4
 
 | | Molybdenum storage protein - P6422, ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ermler, U, Bruenle, S. | | Deposit date: | 2019-04-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molybdate pumping into the molybdenum storage protein via an ATP-powered piercing mechanism.
Proc.Natl.Acad.Sci.USA, 2019
|
|
5O3M
 
 | |
5O3N
 
 | | Crystal structure of E. cloacae 3,4-dihydroxybenzoic acid decarboxylase (AroY) reconstituted with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3,4-dihydroxybenzoate decarboxylase, GLYCEROL, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2017-05-24 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7ZQQ
 
 | | Molybdenum storage protein - LB131H | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ermler, U, Aziz, I. | | Deposit date: | 2022-05-02 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The molybdenum storage protein forms and deposits distinct polynuclear tungsten oxygen aggregates.
J.Inorg.Biochem., 234, 2022
|
|
7ZR4
 
 | |
