3AVU
 
 | | Structure of viral RNA polymerase complex 2 | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVW
 
 | | Structure of viral RNA polymerase complex 4 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVX
 
 | | Structure of viral RNA polymerase complex 5 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVV
 
 | | Structure of viral RNA polymerase complex 3 | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.119 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVY
 
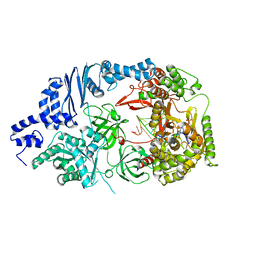 | | Structure of viral RNA polymerase complex 6 | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3D2M
 
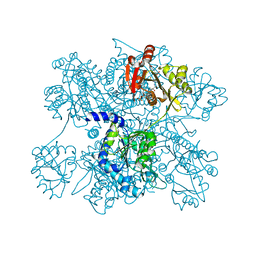 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-glutamate | | Descriptor: | COENZYME A, GLUTAMIC ACID, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
3D2P
 
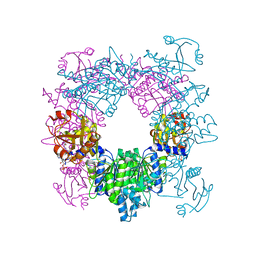 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-arginine | | Descriptor: | ARGININE, COENZYME A, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
4I49
 
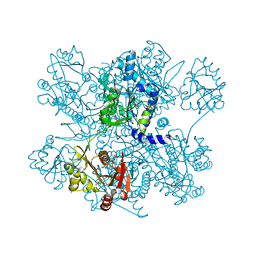 | | Structure of ngNAGS bound with bisubstrate analog CoA-NAG | | Descriptor: | (2S)-2-({(3S,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14,20-trioxo-2,4,6-trioxa-18-thia-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaicosan-20-yl}amino)pentanedioic acid (non-preferred name), Amino-acid acetyltransferase, SULFATE ION | | Authors: | Shi, D, Zhao, G, Allewell, N.M, Tuchman, M. | | Deposit date: | 2012-11-27 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the complex of Neisseria gonorrhoeae N-acetyl-l-glutamate synthase with a bound bisubstrate analog.
Biochem.Biophys.Res.Commun., 430, 2013
|
|
4IWY
 
 | | SeMet-substituted RimK structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Ribosomal protein S6 modification protein, ... | | Authors: | Shi, D, Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of Escherichia coli RimK, an ATP-grasp fold, l-glutamyl ligase enzyme.
Proteins, 81, 2013
|
|
4K30
 
 | | Structure of the N-acetyltransferase domain of human N-acetylglutamate synthase | | Descriptor: | N-ACETYL-L-GLUTAMATE, N-acetylglutamate synthase, mitochondrial | | Authors: | Shi, D, Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Crystal structure of the N-acetyltransferase domain of human N-acetyl-L-glutamate synthase in complex with N-acetyl-L-glutamate provides insights into its catalytic and regulatory mechanisms.
Plos One, 8, 2013
|
|
4IWX
 
 | | Rimk structure at 2.85A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Ribosomal protein S6 modification protein, ... | | Authors: | Shi, D, Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structure and function of Escherichia coli RimK, an ATP-grasp fold, l-glutamyl ligase enzyme.
Proteins, 81, 2013
|
|
6KBR
 
 | |
5ZU2
 
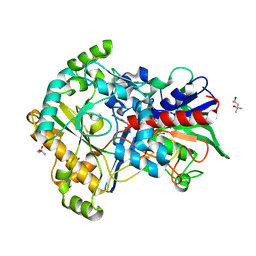 | | Effect of mutation (R554A) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mikami, B, Uchida, H, Doubayashi, D. | | Deposit date: | 2018-05-06 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.441 Å) | | Cite: | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
5ZU3
 
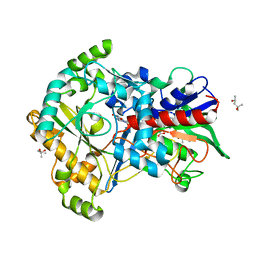 | | Effect of mutation (R554K) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Formate oxidase, ... | | Authors: | Mikami, B, Uchida, H, Doubayashi, D. | | Deposit date: | 2018-05-06 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
1BG7
 
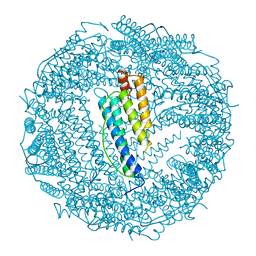 | | LOCALIZED UNFOLDING AT THE JUNCTION OF THREE FERRITIN SUBUNITS. A MECHANISM FOR IRON RELEASE? | | Descriptor: | CALCIUM ION, FERRITIN | | Authors: | Takagi, H, Shi, D, Ha, Y, Allewell, N.M, Theil, E.C. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Localized unfolding at the junction of three ferritin subunits. A mechanism for iron release?
J.Biol.Chem., 273, 1998
|
|
6PQ0
 
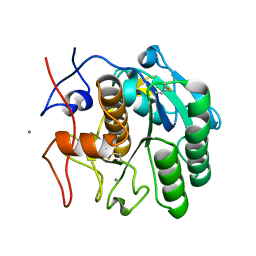 | | LCP-embedded Proteinase K treated with MPD | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
6PQ4
 
 | | LCP-embedded Proteinase K treated with lipase | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
3M4J
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible Post-Translational Carboxylation Modulates the Enzymatic Activity of N-Acetyl-l-ornithine Transcarbamylase.
Biochemistry, 49, 2010
|
|
3M4N
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302A mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
3M5D
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302R mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
3M5C
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302E mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
5K7R
 
 | | MicroED structure of trypsin at 1.7 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7T
 
 | | MicroED structure of thermolysin at 2.5 A resolution | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7Q
 
 | | MicroED structure of thaumatin at 2.5 A resolution | | Descriptor: | Thaumatin-1 | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7N
 
 | | MicroED structure of tau VQIVYK peptide at 1.1 A resolution | | Descriptor: | VQIVYK | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
