1CR1
 
 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTTP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR4
 
 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTDP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
4QX3
 
 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX2
 
 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QX1
 
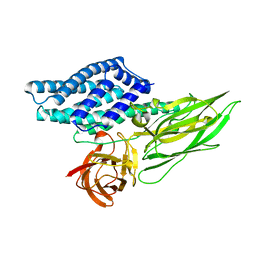 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4DCL
 
 | | Computationally Designed Self-assembling tetrahedron protein, T308, Crystallized in space group F23 | | Descriptor: | Putative acetyltransferase SACOL2570 | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4DDF
 
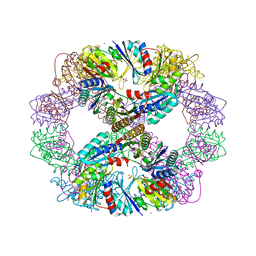 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group P4 | | Descriptor: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
3CJB
 
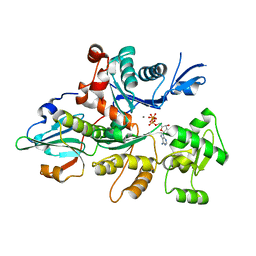 | | Actin dimer cross-linked by V. cholerae MARTX toxin and complexed with Gelsolin-segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sawaya, M.R, Kudryashov, D.S, Pashkov, I, Reisler, E, Yeates, T.O. | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CJH
 
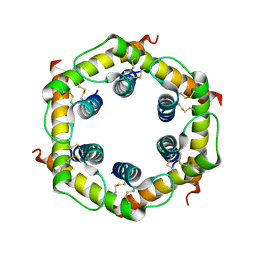 | | Tim8-Tim13 complex | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM13, Mitochondrial import inner membrane translocase subunit TIM8 | | Authors: | Sawaya, M.R, Schmid, E, Beverly, K.N, Koehler, C.M. | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Tim8-Tim13 complex has multiple substrate binding sites and binds cooperatively to Tim23
J.Mol.Biol., 382, 2008
|
|
3CJC
 
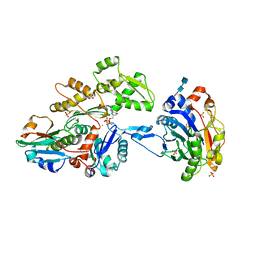 | | Actin dimer cross-linked by V. cholerae MARTX toxin and complexed with DNase I and Gelsolin-segment 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Sawaya, M.R, Kudryashov, D.S, Pashkov, I, Reisler, E, Yeates, T.O. | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4EGG
 
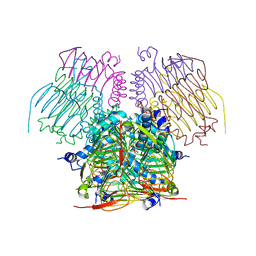 | | Computationally Designed Self-assembling tetrahedron protein, T310 | | Descriptor: | GLYCEROL, Putative acetyltransferase SACOL2570 | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
8DDG
 
 | |
8DDH
 
 | |
8DDF
 
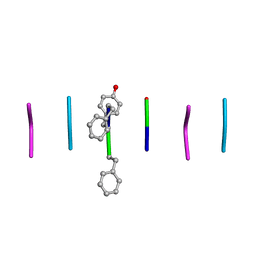 | | Quasi-racemic mixture of L-FWF and D-FYF peptide reveals rippled beta-sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, DPN-DTY-DPN, PHE-TRP-PHE | | Authors: | Sawaya, M.R, Hazari, A, Eisenberg, D.E. | | Deposit date: | 2022-06-18 | | Release date: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The rippled beta-sheet layer configuration-a novel supramolecular architecture based on predictions by Pauling and Corey.
Chem Sci, 13, 2022
|
|
7ROJ
 
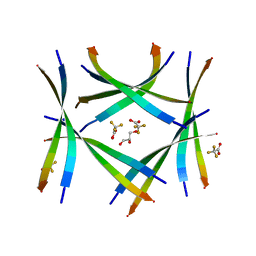 | | Amyloid-related segment of alphaB-crystallin residues 90-100 with G95W mutation | | Descriptor: | Alpha-crystallin B chain peptide, GLYCEROL, SODIUM ION, ... | | Authors: | Sawaya, M.R, Do, T.D, Eisenberg, D.S. | | Deposit date: | 2021-07-30 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic view of an amyloid dodecamer exhibiting selective cellular toxic vulnerability in acute brain slices.
Protein Sci., 31, 2022
|
|
7ROL
 
 | |
6AS9
 
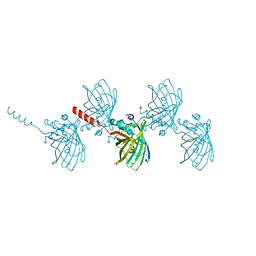 | | Filamentous Assembly of Green Fluorescent Protein Supported by a C-terminal fusion of 18-residues, viewed in space group P212121 form 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Green fluorescent protein | | Authors: | Sawaya, M.R, Heller, D.M, McPartland, L, Hochschild, A, Eisenberg, D.S. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-30 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Atomic insights into the genesis of cellular filaments by globular proteins.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8T84
 
 | | Racemic mixture of amyloid beta segment 35-MVGGVV-40 forms heterochiral rippled beta-sheet, includes hexafluoroisopropanol | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, Racemic mixture of amyloid beta segment 35-MVGGVV-40 | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Racemic Peptides from Amyloid beta and Amylin Form Rippled beta-Sheets Rather Than Pleated beta-Sheets.
J.Am.Chem.Soc., 145, 2023
|
|
8T86
 
 | |
8T82
 
 | | Racemic mixture of amyloid beta segment 35-MVGGVV-40 forms heterochiral rippled beta-sheet, includes pentafluoropropionic acid | | Descriptor: | amyloid beta segment 35-MVGGVV-40, racemic mixture, pentafluoropropanoic acid | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Racemic Peptides from Amyloid beta and Amylin Form Rippled beta-Sheets Rather Than Pleated beta-Sheets.
J.Am.Chem.Soc., 145, 2023
|
|
8T89
 
 | |
5K2F
 
 | | Structure of NNQQNY from yeast prion Sup35 with cadmium acetate determined by MicroED | | Descriptor: | ACETATE ION, CADMIUM ION, Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2E
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate determined by MicroED | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2G
 
 | |
5K2H
 
 | |
