3VQY
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with BocLys and AMPPNP (form 2) | | Descriptor: | MAGNESIUM ION, N~6~-(tert-butoxycarbonyl)-L-lysine, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-02 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VQV
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AMPPNP (re-refined) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyrrolysine--tRNA ligase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VYW
 
 | | Crystal structure of MNMC2 from Aquifex Aeolicus | | Descriptor: | BENZAMIDINE, MNMC2, S-ADENOSYLMETHIONINE | | Authors: | Shibata, R, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-17 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization and structure of the Aquifex aeolicus protein DUF752: a bacterial tRNA-methyltransferase (MnmC2) functioning without the usually fused oxidase domain (MnmC1).
J.Biol.Chem., 287, 2012
|
|
5AVM
 
 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
5B6O
 
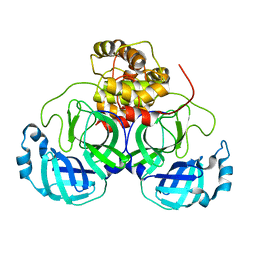 | | Crystal structure of MS8104 | | Descriptor: | 3C-like proteinase | | Authors: | Wang, H, Kim, Y, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
3VNP
 
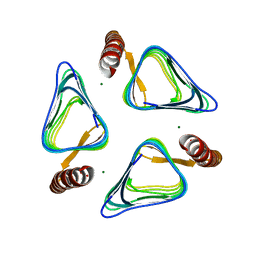 | | Crystal structure of hypothetical protein (GK2848) from Geobacillus Kaustophilus | | Descriptor: | ACETIC ACID, Hypothetical conserved protein, MAGNESIUM ION | | Authors: | Karthe, P.P, Kumarevel, T.S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-01-17 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of hypothetical protein (GK2848) from Geobacillus Kaustophilus
To be Published
|
|
4NYO
 
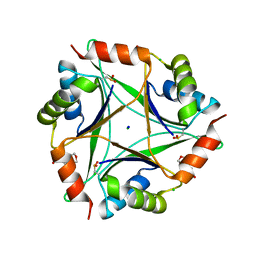 | | The 1.8 Angstrom Crystal Structure of the Periplasmic Divalent Cation Tolerance Protein Cuta from Pyrococcus Horikoshii OT3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Divalent-cation tolerance protein CutA, ... | | Authors: | Bagautdinov, B, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of the CutA1 proteins from Thermus thermophilus and Pyrococcus horikoshii: characterization of metal-binding sites and metal-induced assembly
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
4NYP
 
 | |
3VIU
 
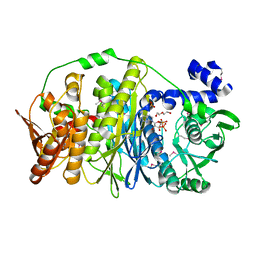 | | Crystal structure of PurL from thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Suzuki, S, Yanai, H, Kanagawa, M, Tamura, S, Watanabe, Y, Fuse, K, Baba, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-10-12 | | Release date: | 2012-01-18 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of N-formylglycinamide ribonucleotide amidotransferase II (PurL) from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1FEX
 
 | |
1GAX
 
 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS VALYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(VAL) AND VALYL-ADENYLATE ANALOGUE | | Descriptor: | N-[VALINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, TRNA(VAL), VALYL-TRNA SYNTHETASE, ... | | Authors: | Fukai, S, Nureki, O, Sekine, S, Shimada, A, Tao, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-06-23 | | Release date: | 2000-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase.
Cell(Cambridge,Mass.), 103, 2000
|
|
1D8J
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1G59
 
 | | GLUTAMYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(GLU). | | Descriptor: | GLUTAMYL-TRNA SYNTHETASE, TRNA(GLU) | | Authors: | Sekine, S, Nureki, O, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-10-31 | | Release date: | 2001-09-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase.
Nat.Struct.Biol., 8, 2001
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | MUTM (FPG) PROTEIN, ZINC ION | | Authors: | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
1FJD
 
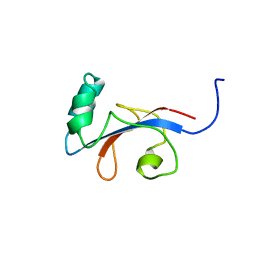 | | HUMAN PARVULIN-LIKE PEPTIDYL PROLYL CIS/TRANS ISOMERASE, HPAR14 | | Descriptor: | PEPTIDYL PROLYL CIS/TRANS ISOMERASE (PPIASE) | | Authors: | Terada, T, Shirouzu, M, Fukumori, Y, Fujimori, F, Ito, Y, Kigawa, T, Yokoyama, S, Uchida, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human parvulin-like peptidyl prolyl cis/trans isomerase, hPar14.
J.Mol.Biol., 305, 2001
|
|
1D9A
 
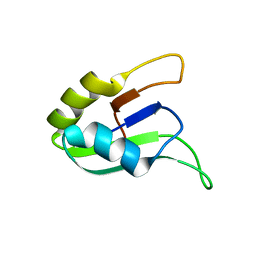 | | SOLUTION STRUCTURE OF THE SECOND RNA-BINDING DOMAIN (RBD2) OF HU ANTIGEN C (HUC) | | Descriptor: | HU ANTIGEN C | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C.
Nucleic Acids Res., 28, 2000
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
1GD8
 
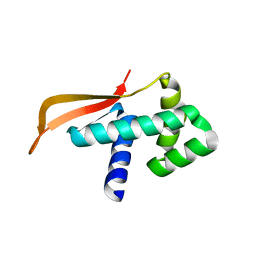 | |
1GD7
 
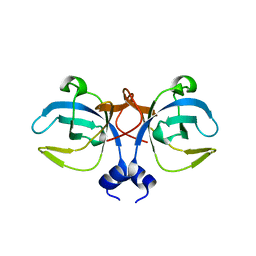 | | CRYSTAL STRUCTURE OF A BIFUNCTIONAL PROTEIN (CSAA) WITH EXPORT-RELATED CHAPERONE AND TRNA-BINDING ACTIVITIES. | | Descriptor: | CSAA PROTEIN | | Authors: | Shibata, T, Inoue, Y, Vassylyev, D.G, Kawaguchi, S, Yokoyama, S, Muller, J, Linde, D, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the ttCsaA protein: an export-related chaperone from Thermus thermophilus.
EMBO J., 20, 2001
|
|
1D8Z
 
 | | SOLUTION STRUCTURE OF THE FIRST RNA-BINDING DOMAIN (RBD1) OF HU ANTIGEN C (HUC) | | Descriptor: | HU ANTIGEN C | | Authors: | Inoue, M, Muto, Y, Sakamoto, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-26 | | Release date: | 2000-04-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies on functional structures of the AU-rich element-binding domains of Hu antigen C.
Nucleic Acids Res., 28, 2000
|
|
1GLN
 
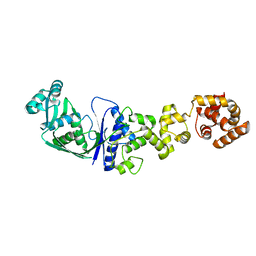 | | ARCHITECTURES OF CLASS-DEFINING AND SPECIFIC DOMAINS OF GLUTAMYL-TRNA SYNTHETASE | | Descriptor: | GLUTAMYL-TRNA SYNTHETASE | | Authors: | Nureki, O, Vassylyev, D.G, Katayanagi, K, Shimizu, T, Sekine, S, Kigawa, T, Miyazawa, T, Yokoyama, S, Morikawa, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1994-07-20 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architectures of class-defining and specific domains of glutamyl-tRNA synthetase.
Science, 267, 1995
|
|
1Q60
 
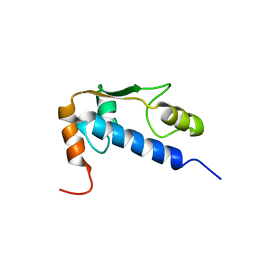 | | Solution Structure of RSGI RUH-004, a GTF2I domain in Mouse cDNA | | Descriptor: | General transcription factor II-I | | Authors: | Doi-Katayama, Y, Hayashi, F, Hirota, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-12 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-004, a GTF2I domain in Mouse cDNA
To be published
|
|
2RS6
 
 | | Solution structure of the N-terminal dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
1NZA
 
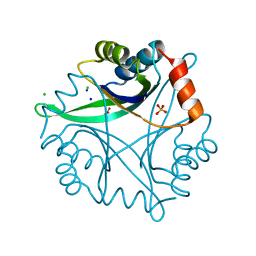 | | Divalent cation tolerance protein (Cut A1) from thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, Divalent cation tolerance protein, GLYCEROL, ... | | Authors: | Bagautdinov, B, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structures of the CutA1 proteins from Thermus thermophilus and Pyrococcus horikoshii: characterization of metal-binding sites and metal-induced assembly.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1NZ8
 
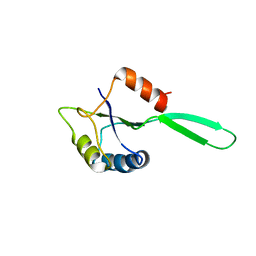 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
