6QWL
 
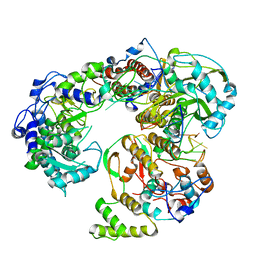 | | Influenza B virus (B/Panama/45) polymerase Hetermotrimer in complex with 3'5' cRNA promoter | | Descriptor: | 3' cRNA, 5' cRNA, Polymerase acidic protein, ... | | Authors: | Keown, J.R, Carrique, L, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
8R3P
 
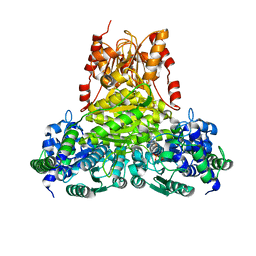 | | Transketolase from Enterococcus faecium in complex with thiamin pyrophosphate | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Ballut, L, Georges, R.N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
8R3Q
 
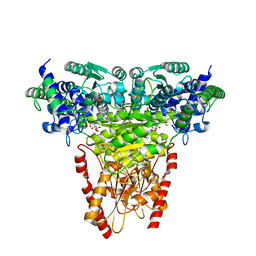 | | Transketolase from Plasmodium falciparum in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Ballut, L, Georges, R.N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
5NL1
 
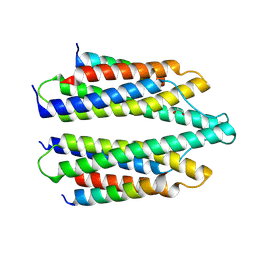 | | Shigella IpaA-VBS3/TBS in complex with the Talin VBS1 domain 488-512 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Invasin IpaA, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Valencia-Gallardo, C, Fontecave, M, Tran Van Nhieu, G. | | Deposit date: | 2017-04-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Shigella IpaA Binding to Talin Stimulates Filopodial Capture and Cell Adhesion.
Cell Rep, 26, 2019
|
|
2XXL
 
 | | Crystal structure of drosophila Grass clip serine protease of Toll pathway | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GRAM-POSITIVE SPECIFIC SERINE PROTEASE, ... | | Authors: | Kellenberger, C, Leone, P, Coquet, L, Jouenne, T, Reichhart, J.M, Roussel, A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Analysis of Grass Clip Serine Protease Involved in Drosophila Toll Pathway Activation.
J.Biol.Chem., 286, 2011
|
|
8R3O
 
 | | Transketolase from Haemophilus influenzae in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Ballut, L, Georges, R.N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
8R3S
 
 | | Transketolase from Staphylococcus aureus in complex with thiamin pyrophosphate | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Ballut, L, Georges, N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
8R3R
 
 | | Transketolase from Streptococcus pneumoniae in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable transketolase, ... | | Authors: | Ballut, L, Georges, R.N, Aghajari, N, Hecquet, L, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-11-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Are Transketolases relevant Targets fighting Human Pathogens? A comparative Biochemical, Bioinformatic and Structural Study
To Be Published
|
|
5FRH
 
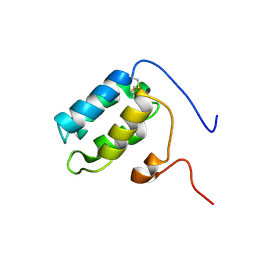 | | Solution structure of oxidised RsrA | | Descriptor: | ANTI-SIGMA FACTOR RSRA | | Authors: | Zdanowski, K, Pecqueur, L, Werner, J, Potts, J.R, Kleanthous, C. | | Deposit date: | 2015-12-17 | | Release date: | 2016-08-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Anti-Sigma Factor Rsra Responds to Oxidative Stress by Reburying its Hydrophobic Core.
Nat.Commun., 7, 2016
|
|
5KH0
 
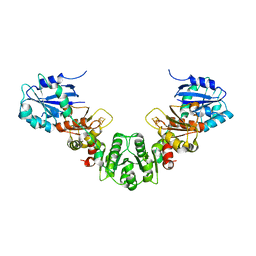 | |
5LAD
 
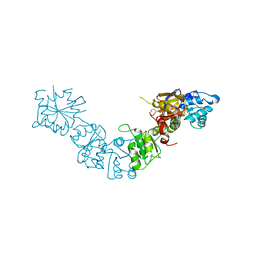 | |
6BNS
 
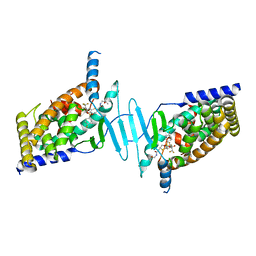 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN BOUND TETHERED WITH SRC co-activator peptide and Compound 25a AKA BICYCLIC HEXAFLUOROISOPROPYL 2 ALCOHOL SULFONAMIDES | | Descriptor: | 2-[(2S)-4-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)-3,4-dihydro-2H-1,4-benzothiazin-2-yl]-N-(2-hydroxy-2-methylpropyl)acetamide, Nuclear receptor subfamily 1 group I member 2,Nuclear receptor coactivator 1 Chimera | | Authors: | DHAR, T.G, GONG, H, WEINSTEIN, D.S, LU, Z, DUAN, J.J.W, STACHURA, S, HAQUE, L, KARMAKAR, A, HEMAGIRI, H, RAUT, D.K, GUPTA, A.K, KHAN, J.A, SACK, J.S, CAMAC, D.M, PUDZIANOWSKI, A.A, WU, D.R, YARDE, M, SHEN, D.R, BOROWSKI, V, XIE, J.H, SUN, H, ARIENZO, C.D, DABROS, M, GALELLA, M.A, WANG, F, WEIGELT, C.A, ZHAO, Q, FOSTER, W, SOMERVILLE, J.E, SALTER-CID, L.M, BARRISH, J.C, CARTER, P.H. | | Deposit date: | 2017-11-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Identification of bicyclic hexafluoroisopropyl alcohol sulfonamides as retinoic acid receptor-related orphan receptor gamma (ROR gamma /RORc) inverse agonists. Employing structure-based drug design to improve pregnane X receptor (PXR) selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5FRF
 
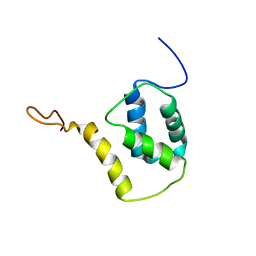 | | Solution structure of reduced and zinc-bound RsrA | | Descriptor: | ANTI-SIGMA FACTOR RSRA, ZINC ION | | Authors: | Zdanowski, K, Pecqueur, L, Werner, J, Potts, J.R, Kleanthous, C. | | Deposit date: | 2015-12-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Anti-Sigma Factor Rsra Responds to Oxidative Stress by Reburying its Hydrophobic Core.
Nat.Commun., 7, 2016
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | Descriptor: | IgG H chain, IgG L chain, Spike glycoprotein | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-15 | | Release date: | 2020-04-29 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6Z97
 
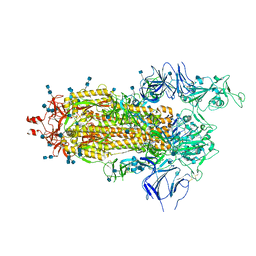 | | Structure of the prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Duyvesteyn, H.M.E, Ren, J, Zhao, Y, Zhou, D, Huo, J, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-06-03 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6Z43
 
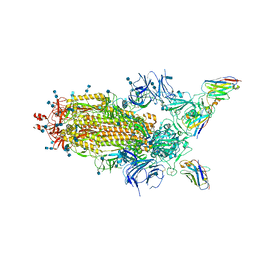 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
6EZA
 
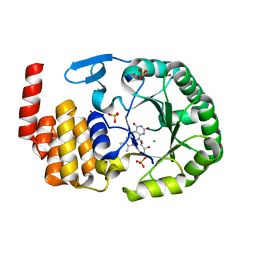 | | Crystal Structure of human tRNA-dihydrouridine(20) synthase catalytic domain E294K mutant | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Bou-Nader, C, Bregeon, D, Pecqueur, L, Vincent, G, Fontecave, M, Hamdane, D. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Electrostatic Potential in the tRNA Binding Evolution of Dihydrouridine Synthases.
Biochemistry, 57, 2018
|
|
6EZB
 
 | | Crystal Structure of human tRNA-dihydrouridine(20) synthase catalytic domain Q305K mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Bou-Nader, C, Bregeon, D, Pecqueur, L, Fontecave, M, Hamdane, D. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Electrostatic Potential in the tRNA Binding Evolution of Dihydrouridine Synthases.
Biochemistry, 57, 2018
|
|
7NHX
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - full transcriptase (Class1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIS
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8192 core | | Descriptor: | Nanobody8192 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-13 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (5.96 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK1
 
 | | 1918 Influenza virus polymerase heterotirmer in complex with vRNA promoters and Nb8201 | | Descriptor: | Nanobody8201, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NKI
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8209 core | | Descriptor: | Nb8209, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NKR
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8210 | | Descriptor: | Nb8210, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-01 | | Last modified: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NHA
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer - Endonuclease and priming loop ordered (Class2a) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIK
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8189 core | | Descriptor: | Nanobody8189 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
