4Z8S
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd)-Native-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZFY
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with ALPHA-METHYL-D-GALACTOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, methyl alpha-D-galactopyranoside, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-22 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZA3
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd)-Native-3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-13 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZFW
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with galactose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZFU
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with N-acetyl D galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4ZLB
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-05-01 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
4X82
 
 | |
8CT4
 
 | | Cryo-EM structure of Mtb Lpd bound to inhibitor complex with 2-((2-cyano-N,5-dimethyl-1H-indole)-7-sulfonamido)-N-(4-(oxetan-3-yl)-3,4-dihydro-2H-benzo[b] [1,4]oxazin-7-yl)acetamide | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, N~2~-(2-cyano-5-methyl-1H-indole-7-sulfonyl)-N~2~-methyl-N-[4-(oxetan-3-yl)-3,4-dihydro-2H-1,4-benzoxazin-7-yl]glycinamide | | Authors: | Kochanczyk, T, Arango, N, Lima, C.D. | | Deposit date: | 2022-05-13 | | Release date: | 2022-05-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Cryo-EM structure of Mtb Lpd bound to the inhibitor 2-((2-cyano-N,5-dimethyl-1H-indole)-7-sulfonamido)-N-(4-(oxetan-3-yl)-3,4-dihydro-2H-benzo[b] [1,4]oxazin-7-yl)acetamide at 2.17 Angstrom resolution
Not published
|
|
3DOF
 
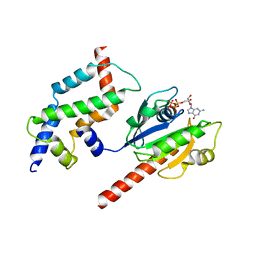 | | Complex of ARL2 and BART, Crystal Form 2 | | Descriptor: | ADP-ribosylation factor-like protein 2, ADP-ribosylation factor-like protein 2-binding protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, T, Li, S, Ding, J. | | Deposit date: | 2008-07-04 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the ARL2-GTP-BART complex reveals a novel recognition and binding mode of small GTPase with effector
Structure, 17, 2009
|
|
6BYX
 
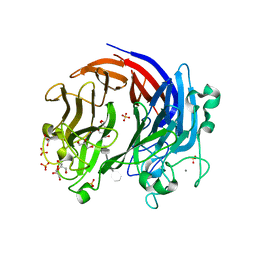 | | Complex structure of LOR107 mutant (R259N) with tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Structure-function analyses of a PL24 family ulvan lyase reveal key features and suggest its catalytic mechanism.
J. Biol. Chem., 293, 2018
|
|
6D2C
 
 | |
6D3U
 
 | | Complex structure of Ulvan lyase from Nonlaben Ulvanivorans- NLR48 | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and functional characterization of PL28 family ulvan lyase NLR48 fromNonlabens ulvanivorans.
J. Biol. Chem., 293, 2018
|
|
8J0G
 
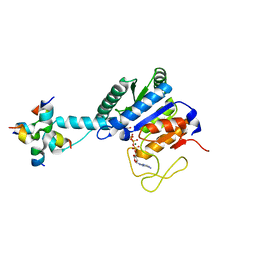 | |
8J28
 
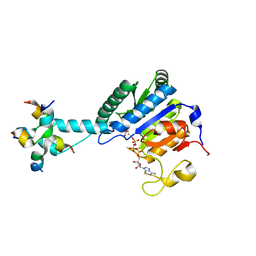 | | GK monomer complexes with G5P and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase B, GAMMA-GLUTAMYL PHOSPHATE | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | GK monomer complexes with G5P and ADP
To Be Published
|
|
8J27
 
 | | GK monomer complexes with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase B | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GK monomer complexes with ADP
To Be Published
|
|
8J0E
 
 | | GK monomer complexes with catalytic intermediate | | Descriptor: | (2~{R})-5-[[[[(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-2-azanyl-5-oxidanylidene-pentanoic acid, Delta-1-pyrroline-5-carboxylate synthase B, MAGNESIUM ION | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | GK monomer complexes with catalytic intermediate
To Be Published
|
|
5Y97
 
 | | Crystal structure of snake gourd seed lectin in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Seed lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Chandran, T, Vijayan, M, Sivaji, N, Surolia, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ligand binding and retention in snake gourd seed lectin (SGSL). A crystallographic, thermodynamic and molecular dynamics study
Glycobiology, 28, 2018
|
|
5Y42
 
 | |
4FTX
 
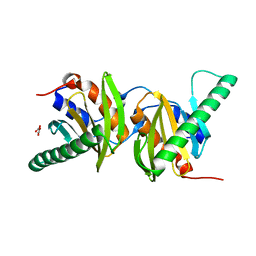 | | Crystal structure of Ego3 homodimer | | Descriptor: | Protein SLM4, SUCCINIC ACID | | Authors: | Zhang, T, Peli-Gulli, M.P, Yang, H, De Virgilio, C, Ding, J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1.
Structure, 20, 2012
|
|
5Y3A
 
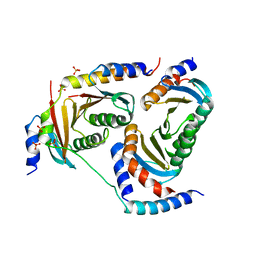 | | Crystal structure of Ragulator complex (p18 49-161) | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1.
Nat Commun, 8, 2017
|
|
4FUW
 
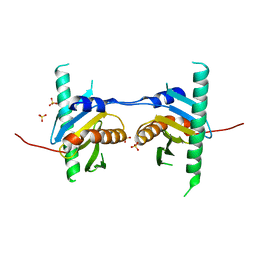 | | Crystal structure of Ego3 mutant | | Descriptor: | Protein SLM4, SULFATE ION | | Authors: | Zhang, T, Peli-Gulli, M.P, Yang, H, De Virgilio, C, Ding, J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1.
Structure, 20, 2012
|
|
5Y38
 
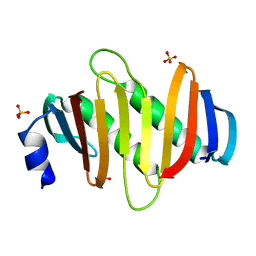 | | Crystal structure of C7orf59-HBXIP complex | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5, SULFATE ION | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1
Nat Commun, 8, 2017
|
|
5Y39
 
 | | Crystal structure of Ragulator complex (p18 76-145) | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1
Nat Commun, 8, 2017
|
|
1X13
 
 | | Crystal structure of E. coli transhydrogenase domain I | | Descriptor: | NAD(P) transhydrogenase subunit alpha | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
1X14
 
 | | Crystal structure of E. coli transhydrogenase domain I with bound NAD | | Descriptor: | NAD(P) transhydrogenase subunit alpha, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
