2OAD
 
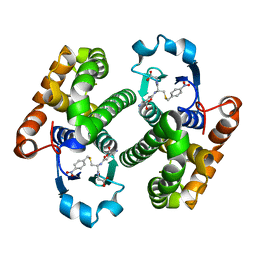 | |
2OAC
 
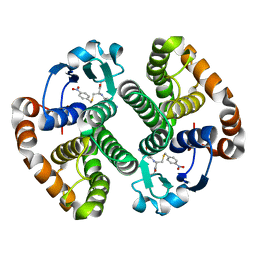 | |
5EY2
 
 | | Crystal structure of CodY from Bacillus cereus | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY | | Authors: | Han, A, Lee, W.C, Son, J, Kim, S.H, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
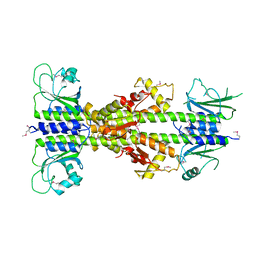
5EY1
 
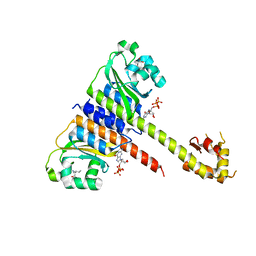 | | Crystal structure of CodY from Staphylococcus aureus with GTP and Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, GUANOSINE-5'-TRIPHOSPHATE, ISOLEUCINE | | Authors: | Han, A, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
5FA9
 
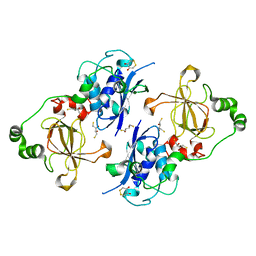 | | Bifunctional Methionine Sulfoxide Reductase AB (MsrAB) from Treponema denticola | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptide methionine sulfoxide reductase MsrA | | Authors: | Han, A, Son, J, Kim, H.-Y, Hwang, K.Y. | | Deposit date: | 2015-12-11 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Essential Role of the Linker Region in the Higher Catalytic Efficiency of a Bifunctional MsrA-MsrB Fusion Protein
Biochemistry, 55, 2016
|
|
5EY0
 
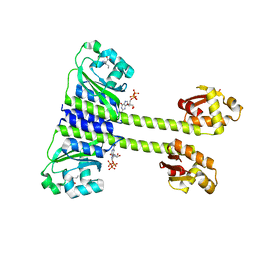 | | Crystal structure of CodY from Staphylococcus aureus with GTP and Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, GUANOSINE-5'-TRIPHOSPHATE, ISOLEUCINE | | Authors: | Han, A, Hwang, K.Y. | | Deposit date: | 2015-11-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism
Nucleic Acids Res., 44, 2016
|
|
5EOS
 
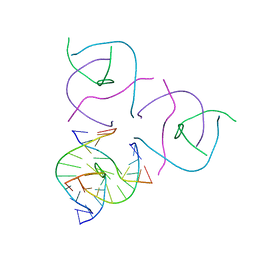 | |
8KB4
 
 | | Cryo-EM structure of human TMEM87A A308M | | Descriptor: | (9R,12R)-15-amino-12-hydroxy-6,12-dioxo-7,11,13-trioxa-12lambda~5~-phosphapentadecan-9-yl undecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 87A,EGFP | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8HSI
 
 | | Cryo-EM structure of human TMEM87A, PE-bound | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8HTT
 
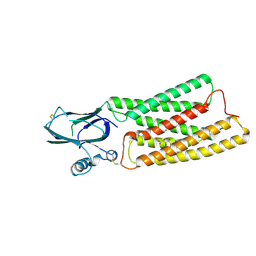 | | Cryo-EM structure of human TMEM87A, gluconate-bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-gluconic acid, Transmembrane protein 87A,EGFP, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
7E72
 
 | | Crystal structure of Tie2-agonistic antibody in complex with human Tie2 Fn2-3 | | Descriptor: | 1,2-ETHANEDIOL, Angiopoietin-1 receptor, the chimeric Fab fragment of 3H7 (heavy chain), ... | | Authors: | Kim, H.M, Jo, G.H, Hong, H.J, Han, A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Structural insights into the clustering and activation of Tie2 receptor mediated by Tie2 agonistic antibody.
Nat Commun, 12, 2021
|
|
8U1R
 
 | |
8H24
 
 | | Leucine-rich alpha-2-glycoprotein 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich alpha-2-glycoprotein, SULFATE ION | | Authors: | Won, S.Y, Park, B.S, Lee, D.S, Kim, H.M, Han, A, Yang, J. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of LRG1 and the functional significance of LRG1 glycan for LPHN2 activation.
Exp.Mol.Med., 55, 2023
|
|
5HXY
 
 | | Crystal structure of XerA recombinase | | Descriptor: | PHOSPHATE ION, Tyrosine recombinase XerA | | Authors: | Hwang, K.Y, Nam, K.H. | | Deposit date: | 2016-01-31 | | Release date: | 2017-02-01 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Thermoplasma acidophilum XerA recombinase shows large C-shape clamp conformation and cis-cleavage mode for nucleophilic tyrosine
FEBS Lett., 590, 2016
|
|
4HKB
 
 | | CH67 Fab (unbound) from the CH65-67 Lineage | | Descriptor: | CH67 heavy chain, CH67 light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-15 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HK3
 
 | | I2 Fab (unbound) from CH65-CH67 Lineage | | Descriptor: | I2 heavy chain, I2 light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HKX
 
 | | Influenza hemagglutinin in complex with CH67 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH67 heavy chain, CH67 light chain, ... | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-15 | | Release date: | 2012-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HK0
 
 | | UCA Fab (unbound) from CH65-CH67 Lineage | | Descriptor: | UCA heavy chain, UCA light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6F7Y
 
 | |
6F7X
 
 | | Crystal structure of dimethylated RSL - cucurbit[7]uril complex, F432 form | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, cucurbit[7]uril, ... | | Authors: | Guagnini, F, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Cucurbit[7]uril-Dimethyllysine Recognition in a Model Protein.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6F7W
 
 | | Crystal structure of dimethylated RSL - cucurbit[7]uril complex, C2221 Form | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SODIUM ION, ... | | Authors: | Guagnini, F, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Cucurbit[7]uril-Dimethyllysine Recognition in a Model Protein.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GL3
 
 | | Crystal structure of human Phosphatidylinositol 4-kinase III beta (PI4KIIIbeta) in complex with ligand 44 | | Descriptor: | (3~{S})-4-(6-azanyl-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl)-~{N}-(4-methoxy-2-methyl-phenyl)-3-methyl-piperazine-1-carboxamide, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Lammens, A, Augustin, M, Steinbacher, S, Reuberson, J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of a Potent, Orally Bioavailable PI4KIII beta Inhibitor (UCB9608) Able To Significantly Prolong Allogeneic Organ Engraftment in Vivo.
J. Med. Chem., 61, 2018
|
|
8GIJ
 
 | |
8DNG
 
 | |
8DNR
 
 | |
