7PGL
 
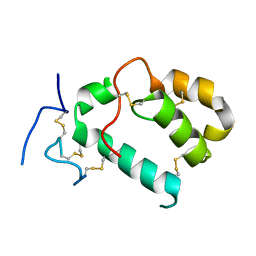 | | HHIP-N, the N-terminal domain of the Hedgehog-Interacting Protein (HHIP), apo-form | | Descriptor: | Hedgehog-interacting protein | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
7PGN
 
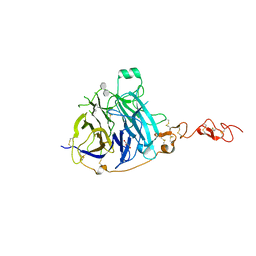 | | HHP-C in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
8ORX
 
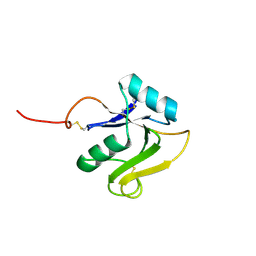 | |
8OS2
 
 | | Structure of the human LYVE-1 (lymphatic vessel endothelial receptor-1) hyaluronan binding domain in an unliganded state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lymphatic vessel endothelial hyaluronic acid receptor 1 | | Authors: | Ni, T, Bannerji, S, Jackson, D.J, Gilbert, R.J.C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A novel sliding interaction with hyaluronan underlies the mode of action of LYVE-1, the receptor for leucocyte entry and trafficking in the lymphatics
To be published
|
|
1OEB
 
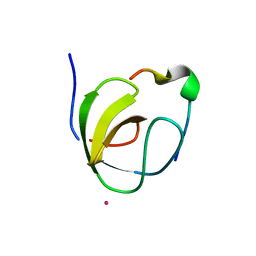 | | Mona/Gads SH3C domain | | Descriptor: | CADMIUM ION, GRB2-RELATED ADAPTOR PROTEIN 2, LYMPHOCYTE CYTOSOLIC PROTEIN 2 | | Authors: | Harkiolaki, M, Lewitzky, M, Gilbert, R.J.C, Jones, E.Y, Bourette, R.P, Mouchiroud, G, Sondermann, H, Moarefi, I, Feller, S.M. | | Deposit date: | 2003-03-24 | | Release date: | 2003-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for SH3 Domain-Mediated High-Affinity Binding between Mona/Gads and Slp-76
Embo J., 22, 2003
|
|
5OWN
 
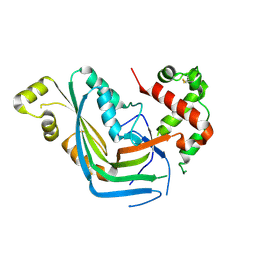 | | Structure of TgPLP1 MACPF domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Perforin-like protein 1 | | Authors: | Ni, T, Gilbert, R.J.C. | | Deposit date: | 2017-09-01 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structures of monomeric and oligomeric forms of theToxoplasma gondiiperforin-like protein 1.
Sci Adv, 4, 2018
|
|
5OUP
 
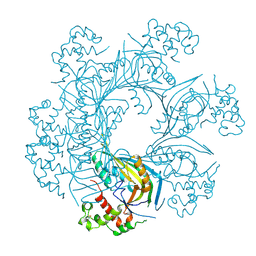 | | Structure of TgPLP1 MACPF domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Perforin-like protein 1 | | Authors: | Ni, T, Gilbert, R.J.C. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of monomeric and oligomeric forms of theToxoplasma gondiiperforin-like protein 1.
Sci Adv, 4, 2018
|
|
5OUO
 
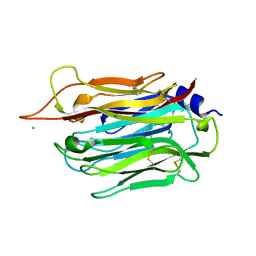 | | Structure of TgPLP1 APCbeta domain | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Perforin-like protein 1 | | Authors: | Ni, T, Gilbert, R.J.C. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structures of monomeric and oligomeric forms of theToxoplasma gondiiperforin-like protein 1.
Sci Adv, 4, 2018
|
|
5OUQ
 
 | | Structure of TgPLP1 MACPF domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Perforin-like protein 1 | | Authors: | Ni, T, Gilbert, R.J.C. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.11 Å) | | Cite: | Structures of monomeric and oligomeric forms of theToxoplasma gondiiperforin-like protein 1.
Sci Adv, 4, 2018
|
|
5OPG
 
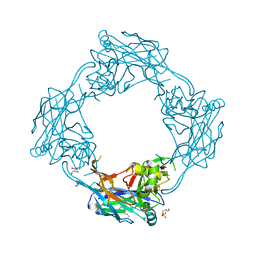 | | Structure of the Hantaan virus Gn glycoprotein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Rissanen, I, Stass, R, Zeltina, A, Li, S, Hepojoki, J, Harlos, K, Gilbert, R.J.C, Huiskonen, J.T, Bowden, T.A. | | Deposit date: | 2017-08-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Transitions of the Conserved and Metastable Hantaviral Glycoprotein Envelope.
J. Virol., 91, 2017
|
|
1YJD
 
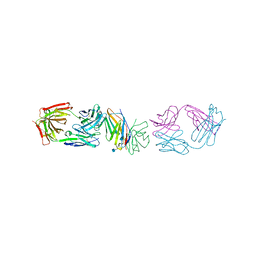 | | Crystal structure of human CD28 in complex with the Fab fragment of a mitogenic antibody (5.11A1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment of 5.11A1 antibody heavy chain, Fab fragment of 5.11A1 antibody light chain, ... | | Authors: | Evans, E.J, Esnouf, R.M, Manso-Sancho, R, Gilbert, R.J.C, James, J.R, Sorensen, P, Stuart, D.I, Davis, S.J. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a soluble CD28-Fab complex
Nat.Immunol., 6, 2005
|
|
3OSK
 
 | | Crystal structure of human CTLA-4 apo homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Yu, C, Sonnen, A.F.-P, Ikemizu, S, Stuart, D.I, Gilbert, R.J.C, Davis, S.J. | | Deposit date: | 2010-09-09 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering
J.Biol.Chem., 286, 2011
|
|
2BK2
 
 | | The prepore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BK1
 
 | | The pore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BPD
 
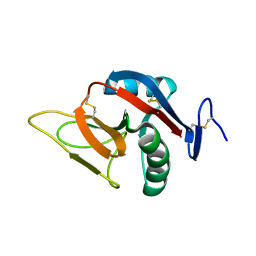 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | DECTIN-1 | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2BPH
 
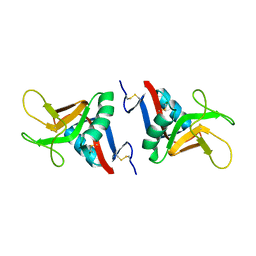 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | DECTIN-1, MAGNESIUM ION | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2BPE
 
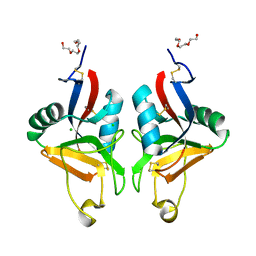 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DECTIN-1, ... | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2CL8
 
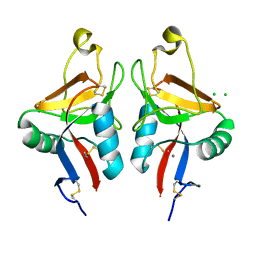 | | Dectin-1 in complex with beta-glucan | | Descriptor: | CALCIUM ION, CHLORIDE ION, DECTIN-1, ... | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2006-04-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2CME
 
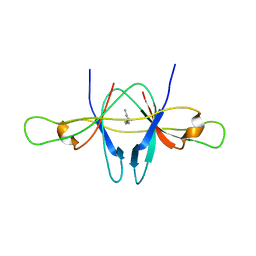 | | The crystal structure of SARS coronavirus ORF-9b protein | | Descriptor: | DECANE, HYPOTHETICAL PROTEIN 5 | | Authors: | Meier, C, Aricescu, A.R, Assenberg, R, Aplin, R.T, Gilbert, R.J.C, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2006-05-06 | | Release date: | 2006-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Orf-9B, a Lipid Binding Protein from the Sars Coronavirus.
Structure, 14, 2006
|
|
2WJW
 
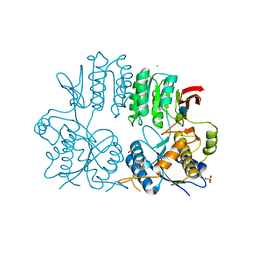 | | Crystal structure of the human ionotropic glutamate receptor GluR2 ATD region at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Clayton, A, Siebold, C, Gilbert, R.J.C, Sutton, G.C, Harlos, K, McIlhinney, R.A.J, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2009-06-01 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Glur2 Amino-Terminal Domain Provides Insights Into the Architecture and Assembly of Ionotropic Glutamate Receptors.
J.Mol.Biol., 392, 2009
|
|
2X44
 
 | | Structure of a strand-swapped dimeric form of CTLA-4 | | Descriptor: | CYTOTOXIC T-LYMPHOCYTE PROTEIN 4 | | Authors: | Sonnen, A.F.-P, Yu, C, Evans, E.J, Stuart, D.I, Davis, S.J, Gilbert, R.J.C. | | Deposit date: | 2010-01-28 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain Metastability: A Molecular Basis for Immunoglobulin Deposition?
J.Mol.Biol., 399, 2010
|
|
2WJX
 
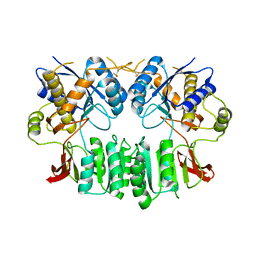 | | Crystal structure of the human ionotropic glutamate receptor GluR2 ATD region at 4.1 A resolution | | Descriptor: | GLUTAMATE RECEPTOR 2 | | Authors: | Clayton, A, Siebold, C, Gilbert, R.J.C, Sutton, G.C, Harlos, K, McIlhinney, R.A.J, Jones, E.Y, Aricescu, A.R. | | Deposit date: | 2009-06-01 | | Release date: | 2009-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal Structure of the Glur2 Amino-Terminal Domain Provides Insights Into the Architecture and Assembly of Ionotropic Glutamate Receptors.
J.Mol.Biol., 392, 2009
|
|
3ZX7
 
 | | Complex of lysenin with phosphocholine | | Descriptor: | LYSENIN, PHOSPHATE ION, PHOSPHOCHOLINE, ... | | Authors: | De Colibus, L, Sonnen, A.F.P, Morris, K.J, Siebert, C.A, Abrusci, P, Plitzko, J, Hodnik, V, Leippe, M, Volpi, E, Anderluh, G, Gilbert, R.J.C. | | Deposit date: | 2011-08-08 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structures of Lysenin Reveal a Shared Evolutionary Origin for Pore-Forming Proteins and its Mode of Sphingomyelin Recognition.
Structure, 20, 2012
|
|
3ZXG
 
 | | lysenin sphingomyelin complex | | Descriptor: | LYSENIN, SULFATE ION, TRIMETHYL-[2-[[(2S,3S)-2-(OCTADECANOYLAMINO)-3-OXIDANYL-BUTOXY]-OXIDANYL-PHOSPHORYL]OXYETHYL]AZANIUM | | Authors: | De Colibus, L, Sonnen, A.F.P, Morris, K.J, Siebert, C.A, Abrusci, P, Plitzko, J, Hodnik, V, Leippe, M, Volpi, E, Anderluh, G, Gilbert, R.J.C. | | Deposit date: | 2011-08-10 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of Lysenin Reveal a Shared Evolutionary Origin for Pore-Forming Proteins and its Mode of Sphingomyelin Recognition.
Structure, 20, 2012
|
|
3ZXD
 
 | | wild-type lysenin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | De Colibus, L, Sonnen, A.F.P, Morris, K.J, Siebert, C.A, Abrusci, P, Plitzko, J, Hodnik, V, Leippe, M, Volpi, E, Anderluh, G, Gilbert, R.J.C. | | Deposit date: | 2011-08-09 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Lysenin Reveal a Shared Evolutionary Origin for Pore-Forming Proteins and its Mode of Sphingomyelin Recognition.
Structure, 20, 2012
|
|
