6B6C
 
 | |
6B6A
 
 | | Beta-Lactamase, 2secs timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6E
 
 | | Beta-Lactamase, mixed with Ceftriaxone, needles crystal form, 500ms | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B68
 
 | | Beta-Lactamase, 100ms timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
7K8H
 
 | | Beta-lactamase mixed with Ceftriaxone, 50ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60006261 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8K
 
 | | Beta-lactamase mixed with Sulbactam, 60ms | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULBACTAM, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8L
 
 | | Beta-lactamase, Unmixed | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8000102 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8F
 
 | | Beta-lactamase mixed with Ceftriaxone, 10ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60003138 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8E
 
 | | Beta-lactamase mixed with Ceftriaxone, 5ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.40005636 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
6ESB
 
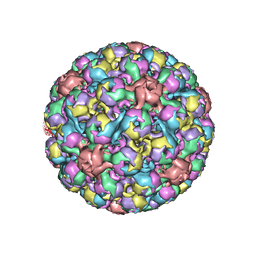 | | BK polyomavirus + 20 mM GT1b oligosaccharide | | Descriptor: | CALCIUM ION, Capsid protein VP1, Minor capsid protein VP2, ... | | Authors: | Hurdiss, D.L, Ranson, N.A. | | Deposit date: | 2017-10-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Structure of an Infectious Human Polyomavirus and Its Interactions with Cellular Receptors.
Structure, 26, 2018
|
|
6P5F
 
 | | Photoactive Yellow Protein PYP Pure Dark | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5E
 
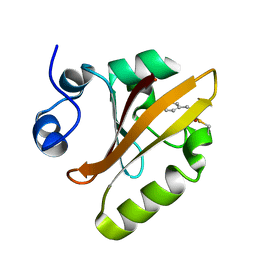 | | Photoactive Yellow Protein PYP 80ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
4ZH1
 
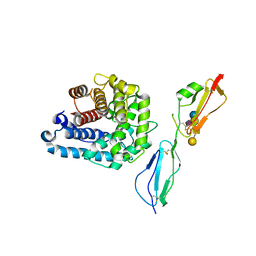 | | Complement factor H in complex with the GM1 glycan | | Descriptor: | Complement C3, Complement factor H, GLYCEROL, ... | | Authors: | Blaum, B.S, Stehle, T. | | Deposit date: | 2015-04-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Complement Factor H and Simian Virus 40 bind the GM1 ganglioside in distinct conformations.
Glycobiology, 26, 2016
|
|
6P5D
 
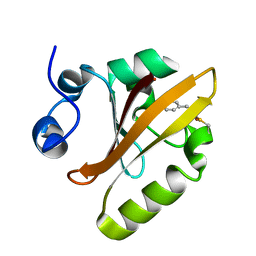 | | Photoactive Yellow Protein PYP 30ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6PGK
 
 | | Membrane Protein Megahertz Crystallography at the European XFEL, Photosystem I XFEL at 2.9 A | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Gisriel, C, Fromme, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Membrane protein megahertz crystallography at the European XFEL.
Nat Commun, 10, 2019
|
|
6P4I
 
 | | Photoactive Yellow Protein PYP 10ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5G
 
 | | Photoactive Yellow Protein PYP Dark Full | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
7TUM
 
 | | Multi-Hit SFX using MHz XFEL sources- first hit | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, SODIUM ION | | Authors: | Darmanin, C, Holmes, S, Abbey, B. | | Deposit date: | 2022-02-03 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
4MJ0
 
 | | BK Polyomavirus VP1 pentamer in complex with GD3 oligosaccharide | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Neu, U, Stroh, L.J, Stehle, T. | | Deposit date: | 2013-09-03 | | Release date: | 2013-11-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus.
Plos Pathog., 9, 2013
|
|
4MJ1
 
 | | unliganded BK Polyomavirus VP1 pentamer | | Descriptor: | CHLORIDE ION, GLYCEROL, VP1 capsid protein | | Authors: | Neu, U, Stroh, L.J, Stehle, T. | | Deposit date: | 2013-09-03 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus.
Plos Pathog., 9, 2013
|
|
3GVL
 
 | | Crystal Structure of endo-neuraminidaseNF | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-beta-neuraminic acid | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3GVK
 
 | | Crystal structure of endo-neuraminidase NF mutant | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3GVJ
 
 | | Crystal structure of an endo-neuraminidaseNF mutant | | Descriptor: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
7ZH2
 
 | | SARS CoV Spike protein, Closed C1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH5
 
 | | SARS CoV Spike protein, Open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
