3CR7
 
 | |
6VFF
 
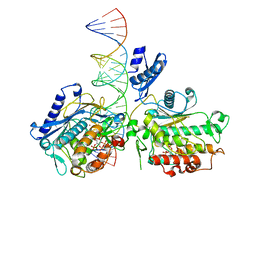 | | Dimer of Human Adenosine Deaminase Acting on dsRNA (ADAR2) mutant E488Q bound to dsRNA sequence derived from human GLI1 gene | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5-R(*GP*CP*UP*CP*GP*CP*GP*AP*UP*GP*CP*UP*(8AZ)P*GP*AP*GP*GP*GP*CP* UP*CP*UP*GP*AP*UP*AP*GP*CP*UP*AP*CP*G)-3), ... | | Authors: | Thuy-boun, A.S, Fisher, A.J, Beal, P.A. | | Deposit date: | 2020-01-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Asymmetric dimerization of adenosine deaminase acting on RNA facilitates substrate recognition.
Nucleic Acids Res., 48, 2020
|
|
3CR8
 
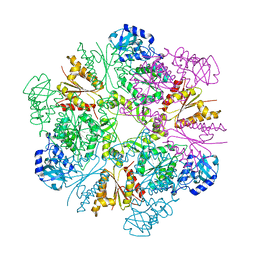 | | Hexameric APS kinase from Thiobacillus denitrificans | | Descriptor: | Sulfate adenylyltransferase, adenylylsulfate kinase | | Authors: | Gay, S.C, Segel, I.H, Fisher, A.J. | | Deposit date: | 2008-04-04 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the two-domain hexameric APS kinase from Thiobacillus denitrificans: structural basis for the absence of ATP sulfurylase activity.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7KFN
 
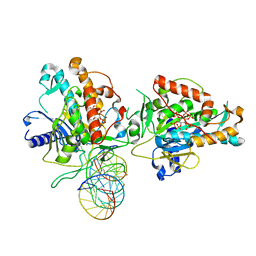 | | Structure of Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA containing a 2'-deoxy Benner's Base Z opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, Gli1 1W5 23mer RNA, Gli1 8AZ 23mer RNA, ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design of RNA Editing Guide Strands: Cytidine Analogs at the Orphan Position.
J.Am.Chem.Soc., 143, 2021
|
|
4R9V
 
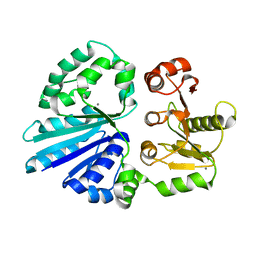 | | Crystal structure of sialyltransferase from photobacterium damselae, residues 113-497 corresponding to the gt-b domain | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Li, Y, Huynh, N, Chen, X, Fisher, A.J. | | Deposit date: | 2014-09-08 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
6XHH
 
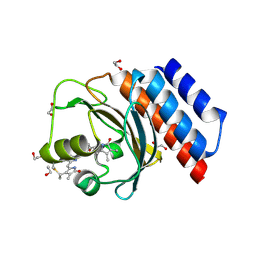 | | Far-red absorbing dark state of JSC1_58120g3 with bound 18-1, 18-2 dihydrobiliverdin IXa (DHBV), the native chromophore precursor | | Descriptor: | 1,2-ETHANEDIOL, JSC1_58120g3, mesobiliverdin IX(alpha) | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XHG
 
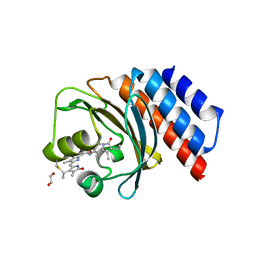 | | Far-red absorbing dark state of JSC1_58120g3 with bound biliverdin IXa (BV) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(~{Z})-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, JSC1_58120g3 | | Authors: | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-10-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8E0F
 
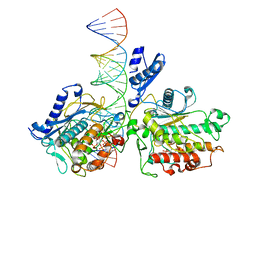 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2-RD) bound to dsRNA containing a G-G pair adjacent to the target site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5-R(*GP*CP*UP*CP*GP*CP*GP*AP*UP*GP*CP*GP*(8AZ)P*GP*AP*GP*GP*GP*CP* UP*CP*UP*GP*AP*UP*AP*GP*CP*UP*AP*CP*G)-3), ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ADAR activation by inducing a syn conformation at guanosine adjacent to an editing site.
Nucleic Acids Res., 50, 2022
|
|
1SFF
 
 | | Structure of gamma-aminobutyrate aminotransferase complex with aminooxyacetate | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
1SZK
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: E211S | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1SZS
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: I50Q | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1SZU
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1SF2
 
 | | Structure of E. coli gamma-aminobutyrate aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
5DJF
 
 | |
5ED2
 
 | |
5ED1
 
 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) mutant E488Q bound to dsRNA sequence derived from S. cerevisiae BDF2 gene | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*CP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1M72
 
 | | Crystal Structure of Caspase-1 from Spodoptera frugiperda | | Descriptor: | 1,2-ETHANEDIOL, Ace-Asp-Glu-Val-Asp-chloromethylketone, Caspase-1 | | Authors: | Forsyth, C.M, Lemongello, D, Friesen, P.D, Fisher, A.J. | | Deposit date: | 2002-07-18 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an invertebrate caspase.
J.Biol.Chem., 279, 2004
|
|
1M0N
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Aminocyclopentanephosphonate | | Descriptor: | 1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]CYCLOPENTYLPHOSPHONIC ACID, 2,2-Dialkylglycine decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1M0P
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Amino-1-phenylethanephosphonate | | Descriptor: | (1R)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]-1-PHENYLETHYLPHOSPHONIC ACID, 2,2-Dialkylglycine Decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1M0Q
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with S-1-aminoethanephosphonate | | Descriptor: | (1S)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]ETHYLPHOSPHONIC ACID, 2,2-Dialkylglycine Decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
4NFO
 
 | | Crystal Structure Analysis of the 16mer GCAGACUUAAGUCUGC | | Descriptor: | GCAGACUUAAGUCUGC, SPERMINE | | Authors: | Beal, P.A, Fisher, A.J, Phelps, K.J, Ibarra-Soza, J.M, Zheng, Y. | | Deposit date: | 2013-10-31 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Click Modification of RNA at Adenosine: Structure and Reactivity of 7-Ethynyl- and 7-Triazolyl-8-aza-7-deazaadenosine in RNA.
Acs Chem.Biol., 9, 2014
|
|
1M7G
 
 | | Crystal structure of APS kinase from Penicillium Chrysogenum: Ternary structure with ADP and APS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE-2',3'-VANADATE, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Lansdon, E.B, Segel, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
1M7H
 
 | | Crystal Structure of APS kinase from Penicillium Chrysogenum: Structure with APS soaked out of one dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-PHOSPHOSULFATE, Adenylylsulfate kinase, ... | | Authors: | Lansdon, E.B, Sege, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
1M8P
 
 | |
1NC2
 
 | | Crystal Structure of Monoclonal Antibody 2D12.5 Fab Complexed with Y-DOTA | | Descriptor: | (S)-2-(4-(2-(2-HYDROXYETHYLTHIO)-ACETAMIDO)-BENZYL)-1,4,7,10-TETRAAZACYCLODODECANE-N,N',N'',N'''-TETRAACETATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Corneillie, T.M, Fisher, A.J, Meares, C.F. | | Deposit date: | 2002-12-04 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two complexes of the rare-earth-DOTA-binding antibody 2D12.5: ligand generality from a chiral system.
J.Am.Chem.Soc., 125, 2003
|
|
