8A1H
 
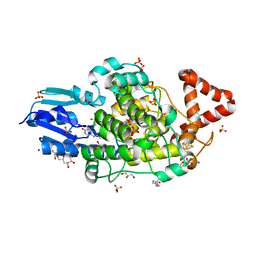 | | Bacterial 6-4 photolyase from Vibrio cholerase | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-4 photolyase (FeS-BCP, ... | | Authors: | Essen, L.-O, Emmerich, H.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Analysis of a Prokaryotic (6-4) Photolyase from the Aquatic Pathogen Vibrio Cholerae † .
Photochem.Photobiol., 99, 2023
|
|
6FN3
 
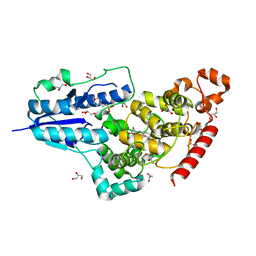 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
6FN0
 
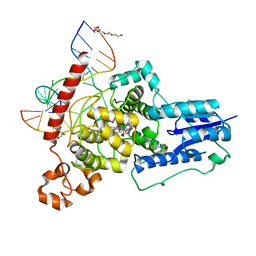 | | The animal-like Cryptochrome from Chlamydomonas reinhardtii in complex with 6-4 DNA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
6FN2
 
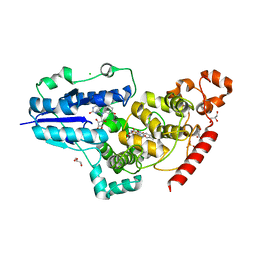 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
6Y98
 
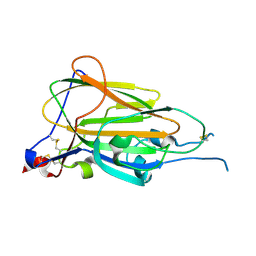 | | Crystal Structure of subtype-switched Epithelial Adhesin 9 to 1 A domain (Epa9-CBL2Epa1) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, PA14 domain-containing protein, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
2GOP
 
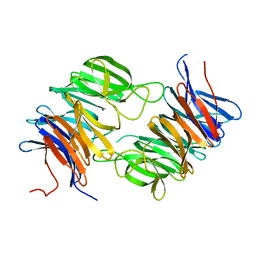 | | The beta-propeller domain of the Trilobed protease from Pyrococcus furiosus reveals an open velcro topology | | Descriptor: | Trilobed Protease | | Authors: | Bosch, J, Tamura, T, Tamura, N, Baumeister, W, Essen, L.-O. | | Deposit date: | 2006-04-13 | | Release date: | 2007-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The beta-propeller domain of the trilobed protease from Pyrococcus furiosus reveals an open Velcro topology.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6Y9J
 
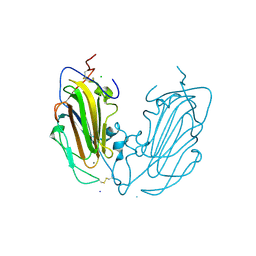 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 9 A domain (Epa1-CBL2Epa9) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epa1p, ... | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
7PZB
 
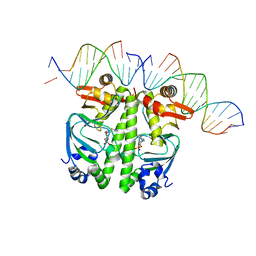 | | Structure of the Clr-cAMP-DNA complex | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*GP*TP*AP*AP*CP*AP*TP*TP*AP*CP*TP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*TP*AP*AP*TP*GP*TP*TP*AP*C)-3'), ... | | Authors: | Werel, L, Essen, L.-O. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Mbio, 14, 2023
|
|
7PZA
 
 | | Structure of the Clr-cAMP-DNA complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*GP*TP*AP*AP*CP*AP*TP*TP*AP*CP*TP*CP*GP)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*TP*AP*AP*TP*GP*TP*TP*AP*C)-3'), ... | | Authors: | Werel, L, Essen, L.-O. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Mbio, 14, 2023
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7QPK
 
 | |
6IAM
 
 | |
5O86
 
 | |
5O8E
 
 | | Mutant of class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ignatz, E, Geisselbrecht, Y, Essen, L.-O. | | Deposit date: | 2017-06-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nicotinamide Adenine Dinucleotides Arrest Photoreduction of Class II DNA Photolyases in FADH ̇ State.
Photochem. Photobiol., 94, 2018
|
|
7AXU
 
 | |
7AXQ
 
 | |
7AXX
 
 | |
7AXP
 
 | |
7AXS
 
 | |
6HOS
 
 | | Structure of the KpFlo2 adhesin domain in complex with glycerol | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, BA75_04148T0, CALCIUM ION, ... | | Authors: | Essen, L.-O, Kock, M, Veelders, M. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of PA14/Flo5-Like Adhesins FromKomagataella pastoris.
Front Microbiol, 9, 2018
|
|
5O8D
 
 | |
4UYS
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | FLOCCULATION PROTEIN FLO11, MAGNESIUM ION, SODIUM ION | | Authors: | Kraushaar, T, Veelders, M, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
4UYR
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | FLOCCULATION PROTEIN FLO11, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Veelders, M, Kraushaar, T, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
4UYT
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, FLOCCULATION PROTEIN FLO11, SODIUM ION | | Authors: | Kraushaar, T, Veelders, M, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
1ASS
 
 | | APICAL DOMAIN OF THE CHAPERONIN FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PHOSPHATE ION, SODIUM ION, THERMOSOME | | Authors: | Klumpp, M, Baumeister, W, Essen, L.-O. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin.
Cell(Cambridge,Mass.), 91, 1997
|
|
