6CJR
 
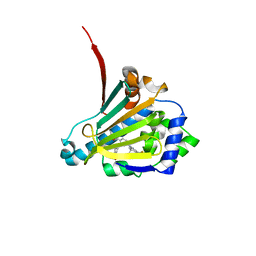 | | Candida albicans Hsp90 nucleotide binding domain in complex with SNX-2112 | | Descriptor: | 1,2-ETHANEDIOL, 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein 90 homolog | | Authors: | Kirkpatrick, M.G, Pizarro, J.C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
6CJJ
 
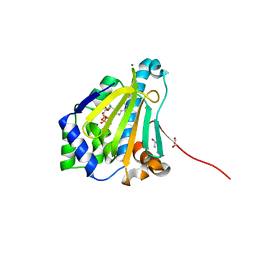 | | Candida albicans Hsp90 nucleotide binding domain in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 90 homolog, ... | | Authors: | Hutchinson, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, STRUCTURAL GENOMICS CONSORTIUM, S.G.C, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
6CJS
 
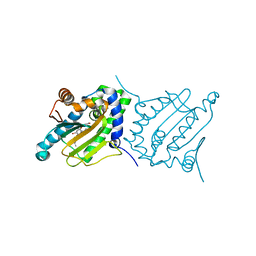 | |
5UZN
 
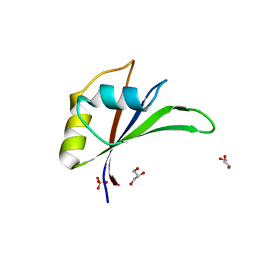 | | Crystal structure of Glorund qRRM3 domain | | Descriptor: | AT27789p, GLYCEROL, SULFATE ION | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
6CJP
 
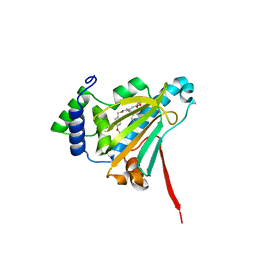 | | Candida albicans Hsp90 nucleotide binding domain in complex with radicicol | | Descriptor: | (1aR,2Z,4E,6E,14R,15aR)-9,11-dihydroxy-6-{[(4-methoxyphenyl)methoxy]imino}-14-methyl-1a,6,7,14,15,15a-hexahydro-12H-oxireno[e][2]benzoxacyclotetradecin-12-one, Heat shock protein 90 homolog | | Authors: | Nation, C, Pizarro, J.C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
6CJL
 
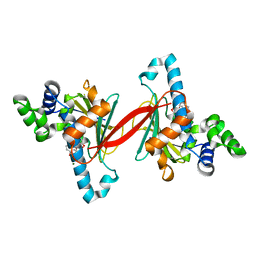 | |
5UZM
 
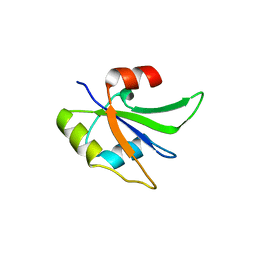 | | Crystal structure of Glorund qRRM2 domain | | Descriptor: | AT27789p | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
6CJI
 
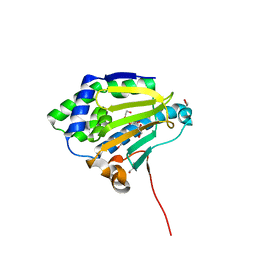 | | Candida albicans Hsp90 nucleotide binding domain | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein 90 homolog | | Authors: | Hutchinson, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, STRUCTURAL GENOMICS CONSORTIUM, S.G.C, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
5UZG
 
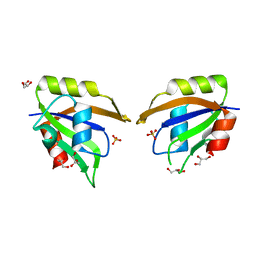 | | Crystal structure of Glorund qRRM1 domain | | Descriptor: | AT27789p, GLYCEROL, SULFATE ION | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
8THN
 
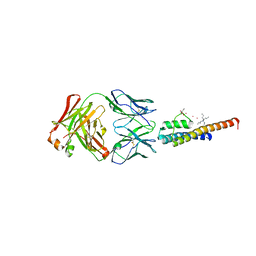 | | KcsA M96V mutant with Y78ester in High K+ | | Descriptor: | KcsA Fab Heavy Chain, KcsA Fab Light Chain, POTASSIUM ION, ... | | Authors: | Reddi, R, Valiyaveetil, F.I. | | Deposit date: | 2023-07-17 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A facile approach for incorporating tyrosine esters to probe ion-binding sites and backbone hydrogen bonds.
J.Biol.Chem., 300, 2023
|
|
2LQ2
 
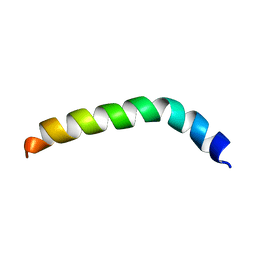 | | Solution structure of de novo designed peptide 4m | | Descriptor: | de novo designed antifreeze peptide 4m | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
2LQ1
 
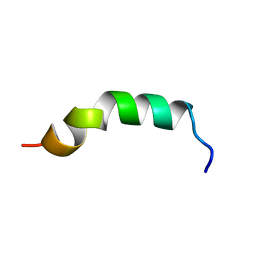 | | Solution structure of de novo designed antifreeze peptide 3 | | Descriptor: | de novo designed antifreeze peptide 3 | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
2LQ0
 
 | | Solution structure of de novo designed antifreeze peptide 1m | | Descriptor: | de novo designed antifreeze peptide 1m | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
