4C34
 
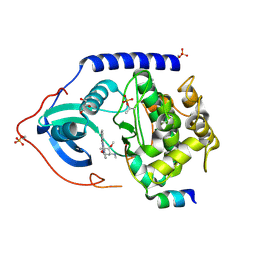 | | PKA-S6K1 Chimera with Staurosporine bound | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, GLYCEROL, ... | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
1OLL
 
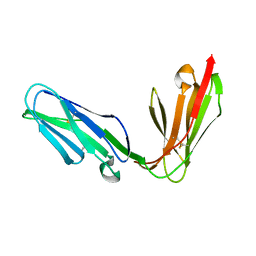 | | Extracellular region of the human receptor NKp46 | | Descriptor: | 1,2-ETHANEDIOL, NK RECEPTOR | | Authors: | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Pesce, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-08-07 | | Release date: | 2003-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the Human Nk Cell Triggering Receptor Nkp46 Ectodomain
Biochem.Biophys.Res.Commun., 309, 2003
|
|
2M2Q
 
 | | Solution structure of MCh-1: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-1 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
2JST
 
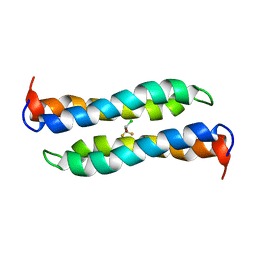 | | Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets II: Halothane Effects on Structure and Dynamics | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, Four-Alpha-Helix Bundle | | Authors: | Cui, T, Bondarenko, V, Ma, D, Canlas, C, Brandon, N.R, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2007-07-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part II: halothane effects on structure and dynamics
Biophys.J., 94, 2008
|
|
4CFX
 
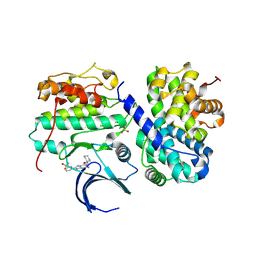 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]benzenesulfonamide, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
2M2R
 
 | | Solution structure of MCh-2: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-2 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
4CFN
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-(cyclohexylmethoxy)-8-(trifluoromethyl)-9H-purin-2-amine, CYCLIN-A2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFV
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methylphenol, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFM
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 6-(cyclohexylmethoxy)-8-(2-methylphenyl)-9H-purin-2-amine, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFU
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-azanyl-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methyl-benzoic acid, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4CFW
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methylbenzenesulfonamide, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4BDB
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]benzene-1,3-diol, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDI
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 1-acetyl-N-(5-methylpyridin-2-yl)piperidine-4-carboxamide, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDA
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydro[1,2,4]triazolo[1,5-a][3,1]benzimidazol-9-ium, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDK
 
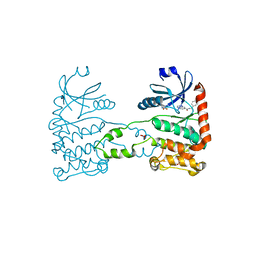 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, CHECKPOINT KINASE 2, N-[(4-methoxyphenyl)methyl]quinoxaline-6-carboxamide, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDG
 
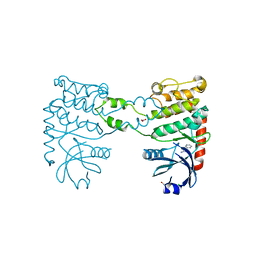 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 3-(PYRIDIN-3-YL)-1H-PYRAZOL-5-AMINE, CHLORIDE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDF
 
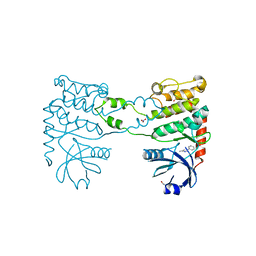 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 5-METHYL-3-PHENYL-1H-PYRAZOLE, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
1Z5L
 
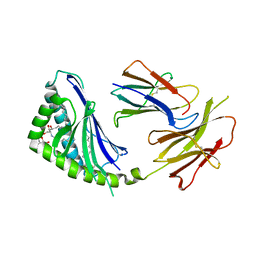 | | Structure of a highly potent short-chain galactosyl ceramide agonist bound to CD1D | | Descriptor: | (2S,3S,4R)-N-OCTANOYL-1-[(ALPHA-D-GALACTOPYRANOSYL)OXY]-2-AMINO-OCTADECANE-3,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Cantu, C, Mattner, J, Zhou, D, Savage, P.B, Bendelac, A, Wilson, I.A, Teyton, L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor.
Nat.Immunol., 6, 2005
|
|
4BDD
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 1-(TERT-BUTYL)-3-(QUINOXALIN-6-YL)UREA, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDC
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-(furan-2-ylmethyl)quinoxaline-6-carboxamide, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BDE
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 6-METHYLQUINAZOLIN-4-AMINE, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4C4E
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, N-(3,4-dimethoxyphenyl)-2-(1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridin-6-amine, ... | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
4C4F
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
4C4H
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, tert-butyl 6-((2-chloro-4-(dimethylcarbamoyl)phenyl)amino)-2-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridine-1-carboxylate | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
4C4G
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
