4F3F
 
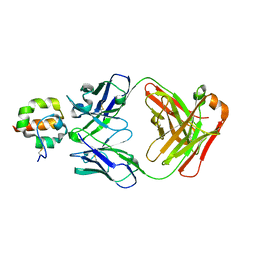 | | Crystal Structure of Msln7-64 MORAb-009 FAB complex | | Descriptor: | MORAb-009 Fab heavy chain, MORAb-009 Fab light chain, Mesothelin | | Authors: | Xia, D, Pastan, I, Ma, J, Tang, W.K, Esser, L. | | Deposit date: | 2012-05-09 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Recognition of mesothelin by the therapeutic antibody MORAb-009: structural and mechanistic insights.
J.Biol.Chem., 287, 2012
|
|
1R6B
 
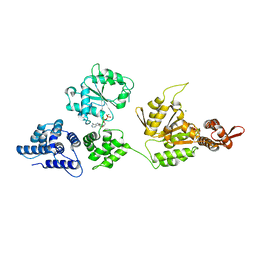 | | High resolution crystal structure of ClpA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ClpA protein, MAGNESIUM ION | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone.
J.Struct.Biol., 146, 2004
|
|
1MBX
 
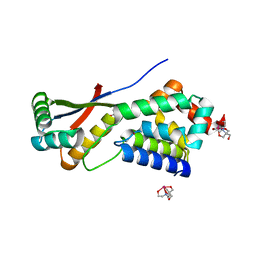 | | CRYSTAL STRUCTURE ANALYSIS OF ClpSN WITH TRANSITION METAL ION BOUND | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, CHLORIDE ION, ... | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of the AAA+ Chaperone, ClpA
J.Biol.Chem., 277, 2002
|
|
1KSF
 
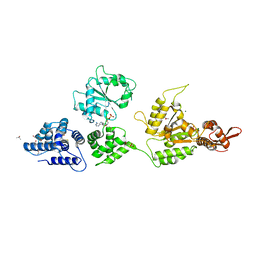 | | Crystal Structure of ClpA, an HSP100 chaperone and regulator of ClpAP protease: Structural basis of differences in Function of the Two AAA+ ATPase domains | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT CLP PROTEASE ATP-BINDING SUBUNIT CLPA, ISOPROPYL ALCOHOL, ... | | Authors: | Guo, F, Maurizi, M.R, Esser, L, Xia, D. | | Deposit date: | 2002-01-12 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of ClpA, an HSP100 chaperone and regulator of ClpAP protease
J.Biol.Chem., 277, 2002
|
|
1MBV
 
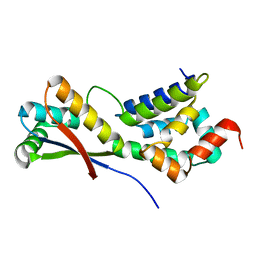 | | CRYSTAL STRUCTURE ANALYSIS OF ClpSN HETERODIMER TETRAGONAL FORM | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, Protein yljA | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of AAA+ Chaperone ClpA
J.Biol.Chem., 277, 2002
|
|
1MBU
 
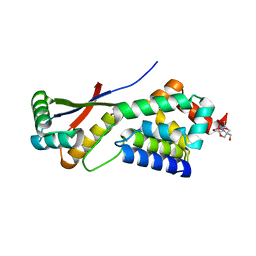 | | Crystal Structure Analysis of ClpSN heterodimer | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, CHLORIDE ION, ... | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of the AAA+ Chaperone, ClpA
J.Biol.Chem., 277, 2002
|
|
1KMP
 
 | | Crystal structure of the Outer Membrane Transporter FecA Complexed with Ferric Citrate | | Descriptor: | CITRIC ACID, FE (III) ION, IRON(III) DICITRATE TRANSPORT PROTEIN FECA, ... | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
1KMO
 
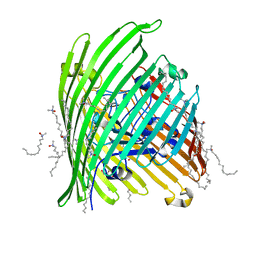 | | Crystal structure of the Outer Membrane Transporter FecA | | Descriptor: | HEPTANE-1,2,3-TRIOL, Iron(III) dicitrate transport protein fecA, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
1R6O
 
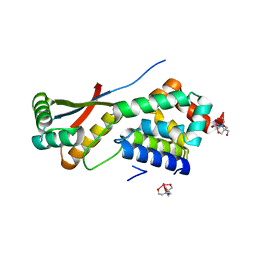 | | ATP-dependent Clp protease ATP-binding subunit clpA/ATP-dependent Clp protease adaptor protein clpS | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA, ATP-dependent Clp protease adaptor protein clpS, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, ... | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the
ClpA chaperone
J.Struct.Biol., 146, 2004
|
|
1R6C
 
 | | High resolution structure of ClpN | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the
ClpA chaperone
J.Struct.Biol., 146, 2004
|
|
1R6Q
 
 | | ClpNS with fragments | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA, ATP-dependent Clp protease adaptor protein clpS, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, ... | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-16 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone.
J.Struct.Biol., 146, 2004
|
|
2VUL
 
 | | Thermostable mutant of ENVIRONMENTALLY ISOLATED GH11 XYLANASE | | Descriptor: | DODECAETHYLENE GLYCOL, GH11 XYLANASE, SULFATE ION | | Authors: | Dumon, C, Varvak, A, Wall, M.A, Flint, J.E, Lewis, R.J, Lakey, J.H, Luginbuhl, P, Healey, S, Todaro, T, Desantis, G, Sun, M, Parra-Gessert, L, Tan, X, Weiner, D.P, Gilbert, H.J. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering Hyperthermostability Into a Gh11 Xylanase is Mediated by Subtle Changes to Protein Structure.
J.Biol.Chem., 283, 2008
|
|
2B4L
 
 | | Crystal structure of the binding protein OpuAC in complex with glycine betaine | | Descriptor: | 1,2-ETHANEDIOL, Glycine betaine-binding protein, TRIMETHYL GLYCINE | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
2B4M
 
 | | Crystal structure of the binding protein OpuAC in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding protein | | Authors: | Horn, C, Sohn-Boesser, L, Breed, J, Welte, W, Schmitt, L, Bremer, E. | | Deposit date: | 2005-09-26 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Determinants for Substrate Specificity of the Ligand-binding Protein OpuAC from Bacillus subtilis for the Compatible Solutes Glycine Betaine and Proline Betaine.
J.Mol.Biol., 357, 2006
|
|
2VUJ
 
 | | Environmentally isolated GH11 xylanase | | Descriptor: | GH11 XYLANASE, GLYCEROL | | Authors: | Dumon, C, Varvak, A, Wall, M.A, Flint, J.E, Lewis, R.J, Lakey, J.H, Luginbuhl, P, Healey, S, Todaro, T, DeSantis, G, Sun, M, Parra-Gessert, L, Tan, X, Weiner, D.P, Gilbert, H.J. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering Hyperthermostability Into a Gh11 Xylanase is Mediated by Subtle Changes to Protein Structure.
J.Biol.Chem., 283, 2008
|
|
1I8E
 
 | | NMR ENSEMBLE OF ION-SELECTIVE LIGAND A22 FOR PLATELET INTEGRIN ALPHAIIB-BETA3 | | Descriptor: | ION-SELECTIVE LIGAND A22 | | Authors: | Smith, J.W, Le Calvez, H, Parra-Gessert, L, Preece, N.E, Jia, X, Assa-Munt, N. | | Deposit date: | 2001-03-13 | | Release date: | 2002-07-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Selection and structure of ion-selective ligands for platelet integrin alpha IIb(beta) 3.
J.Biol.Chem., 277, 2002
|
|
1I6Y
 
 | | NMR ENSEMBLE OF ION-SELECTIVE LIGAND A1 FOR PLATELET INTEGRIN ALPHAIIB-BETA3 | | Descriptor: | ION-SELECTIVE LIGAND A1 | | Authors: | Smith, J.W, Le Calvez, H, Parra-Gessert, L, Preece, N.E, Jia, X, Assa-Munt, N. | | Deposit date: | 2001-03-06 | | Release date: | 2002-07-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Selection and structure of ion-selective ligands for platelet integrin alpha IIb(beta) 3.
J.Biol.Chem., 277, 2002
|
|
1I93
 
 | | NMR ENSEMBLE OF ION-SELECTIVE LIGAND D16 FOR PLATELET INTEGRIN ALPHAIIB-BETA3 | | Descriptor: | ION-SELECTIVE LIGAND D16 | | Authors: | Smith, J.W, Le Calvez, H, Parra-Gessert, L, Preece, N.E, Jia, X, Assa-Munt, N. | | Deposit date: | 2001-03-17 | | Release date: | 2002-07-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Selection and structure of ion-selective ligands for platelet integrin alpha IIb(beta) 3.
J.Biol.Chem., 277, 2002
|
|
1I98
 
 | | NMR ENSEMBLE OF ION-SELECTIVE LIGAND D18 FOR PLATELET INTEGRIN ALPHAIIB-BETA3 | | Descriptor: | ION-SELECTIVE LIGAND D18 | | Authors: | Smith, J.W, Le Calvez, H, Parra-Gessert, L, Preece, N.E, Jia, X, Assa-Munt, N. | | Deposit date: | 2001-03-18 | | Release date: | 2002-07-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Selection and structure of ion-selective ligands for platelet integrin alpha IIb(beta) 3.
J.Biol.Chem., 277, 2002
|
|
6NHH
 
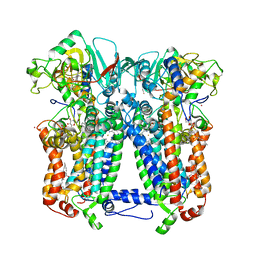 | | Rhodobacter sphaeroides bc1 with azoxystrobin | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Cytochrome b, Cytochrome c1, ... | | Authors: | Xia, D, Zhou, F, Yu, C.A. | | Deposit date: | 2018-12-21 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacterial cytochromebc1in complex with azoxystrobin reveals a conformational switch of the Rieske iron-sulfur protein subunit.
J.Biol.Chem., 294, 2019
|
|
7NAB
 
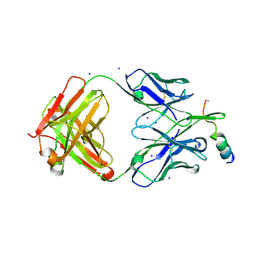 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | Descriptor: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-06-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
3HU3
 
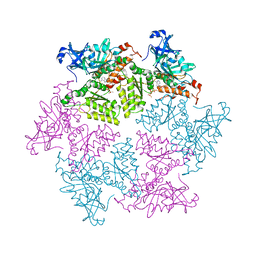 | | Structure of p97 N-D1 R155H mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.-K. | | Deposit date: | 2009-06-12 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
3HU1
 
 | | Structure of p97 N-D1 R95G mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.-K. | | Deposit date: | 2009-06-12 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
3HU2
 
 | | Structure of p97 N-D1 R86A mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.-K. | | Deposit date: | 2009-06-12 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
6U1Y
 
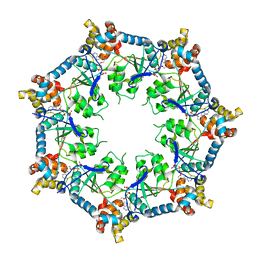 | | bcs1 AAA domain | | Descriptor: | MAGNESIUM ION, Mitochondrial chaperone BCS1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2019-08-17 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
