2GKG
 
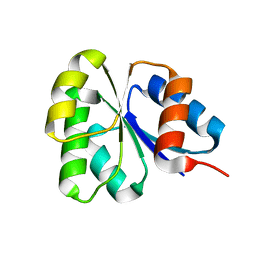 | | Receiver domain from Myxococcus xanthus social motility protein FrzS | | Descriptor: | response regulator homolog | | Authors: | Echols, N, Fraser, J, Merlie, J, Zusman, D, Alber, T. | | Deposit date: | 2006-04-01 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
4PPR
 
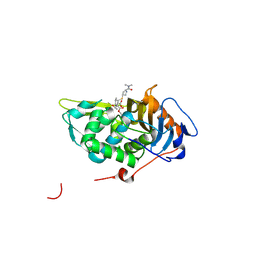 | | Crystal structure of Mycobacterium tuberculosis D,D-peptidase Rv3330 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein DacB1 | | Authors: | Prigozhin, D.M, Huizar, J.P, Mavrici, D, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-27 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-specific adaptations in the structures of two penicillin-binding proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
3M4U
 
 | |
4G1H
 
 | |
4G1J
 
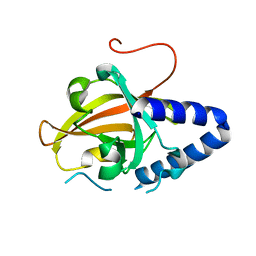 | | Sortase C1 of GBS Pilus Island 1 | | Descriptor: | Sortase family protein | | Authors: | Cozzi, R, Prigozhin, D.M, Alber, T. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for group B streptococcus pilus 1 sortases C regulation and specificity.
Plos One, 7, 2012
|
|
4ESQ
 
 | | Crystal structure of the extracellular domain of PknH from Mycobacterium tuberculosis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Serine/threonine protein kinase, TERBIUM(III) ION | | Authors: | Cavazos, A, Prigozhin, D.M, Alber, T. | | Deposit date: | 2012-04-23 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the sensor domain of Mycobacterium tuberculosis PknH receptor kinase reveals a conserved binding cleft.
J.Mol.Biol., 422, 2012
|
|
2H34
 
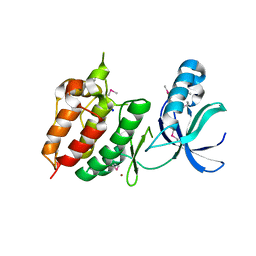 | | Apoenzyme crystal structure of the tuberculosis serine/threonine kinase, PknE | | Descriptor: | BROMIDE ION, SODIUM ION, Serine/threonine-protein kinase pknE | | Authors: | Gay, L.M, Ng, H.L, Alber, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conserved Dimer and Global Conformational Changes in the Structure of apo-PknE Ser/Thr Protein Kinase from Mycobacterium tuberculosis.
J.Mol.Biol., 360, 2006
|
|
1L10
 
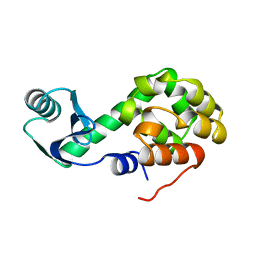 | |
1DCP
 
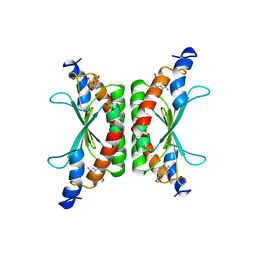 | | DCOH, A BIFUNCTIONAL PROTEIN-BINDING TRANSCRIPTIONAL COACTIVATOR, COMPLEXED WITH BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, DCOH | | Authors: | Cronk, J.D, Endrizzi, J.A, Alber, T. | | Deposit date: | 1996-05-16 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution structures of the bifunctional enzyme and transcriptional coactivator DCoH and its complex with a product analogue.
Protein Sci., 5, 1996
|
|
3N6Q
 
 | | Crystal structure of YghZ from E. coli | | Descriptor: | MAGNESIUM ION, YghZ aldo-keto reductase | | Authors: | Zubieta, C, Totir, M, Echols, N, May, A, Alber, T. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
1G39
 
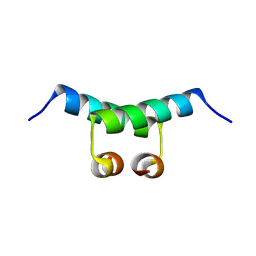 | | WILD-TYPE HNF-1ALPHA DIMERIZATION DOMAIN | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Endrizzi, J.A, Cronk, J.D, Holton, J, Alber, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | High-resolution structure of the HNF-1alpha dimerization domain.
Biochemistry, 39, 2000
|
|
1G2Y
 
 | | HNF-1ALPHA DIMERIZATION DOMAIN, WITH SELENOMETHIONINE SUBSTITUED AT LEU 12 | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Endrizzi, J.A, Cronk, J.D, Holton, J, Alber, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution structure of the HNF-1alpha dimerization domain.
Biochemistry, 39, 2000
|
|
1DCO
 
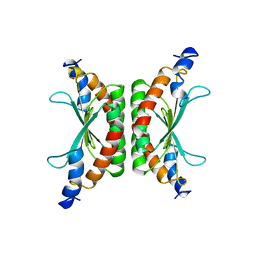 | |
1G2Z
 
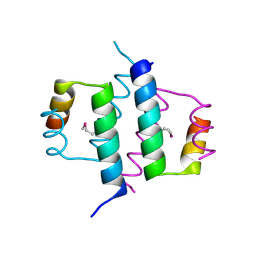 | | DIMERIZATION DOMAIN OF HNF-1ALPHA WITH A LEU 13 SELENOMETHIONINE SUBSTITUTION | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Endrizzi, J.A, Cronk, J.D, Holton, J, Alber, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-resolution structure of the HNF-1alpha dimerization domain.
Biochemistry, 39, 2000
|
|
1RU0
 
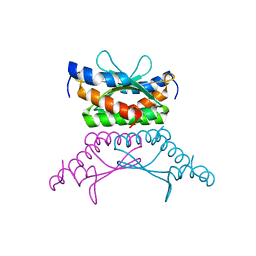 | | Crystal structure of DCoH2, a paralog of DCoH, the Dimerization Cofactor of HNF-1 | | Descriptor: | DcoH-like protein DCoHm | | Authors: | Rose, R.B, Pullen, K.E, Bayle, J.H, Crabtree, G.R, Alber, T. | | Deposit date: | 2003-12-10 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and structural basis for partially redundant enzymatic and transcriptional functions of DCoH and DCoH2
Biochemistry, 43, 2004
|
|
1TGG
 
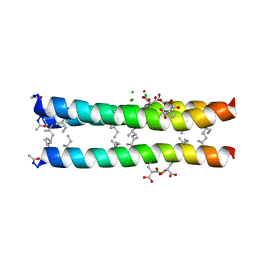 | | RH3 DESIGNED RIGHT-HANDED COILED COIL TRIMER | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, right-handed coiled coil trimer | | Authors: | Plecs, J.J, Harbury, P.B, Kim, P.S, Alber, T. | | Deposit date: | 2004-05-28 | | Release date: | 2004-10-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural test of the parameterized-backbone method for protein design
J.Mol.Biol., 342, 2004
|
|
2NT4
 
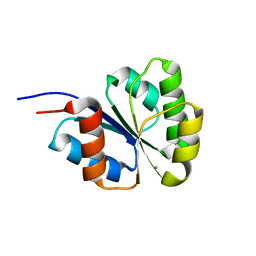 | | Receiver domain from Myxococcus xanthus social motility protein FrzS (H92F mutant) | | Descriptor: | CHLORIDE ION, Response regulator homolog | | Authors: | Echols, N, Fraser, J, Weisfield, S, Merlie, J, Zusman, D, Alber, T. | | Deposit date: | 2006-11-06 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
3KA4
 
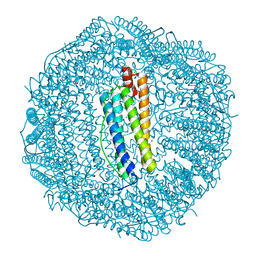 | | Frog M-ferritin with cobalt | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferritin, ... | | Authors: | Tosha, T, Ng, H.L, Theil, E, Alber, T, Bhattasali, O. | | Deposit date: | 2009-10-18 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Moving Metal Ions through Ferritin-Protein Nanocages from Three-Fold Pores to Catalytic Sites.
J.Am.Chem.Soc., 132, 2010
|
|
3KA6
 
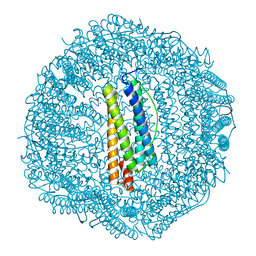 | | Frog M-ferritin, EED mutant, with cobalt | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferritin, ... | | Authors: | Tosha, T, Ng, H.L, Theil, E, Alber, T, Bhattasali, O. | | Deposit date: | 2009-10-18 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Moving Metal Ions through Ferritin-Protein Nanocages from Three-Fold Pores to Catalytic Sites.
J.Am.Chem.Soc., 132, 2010
|
|
1I1J
 
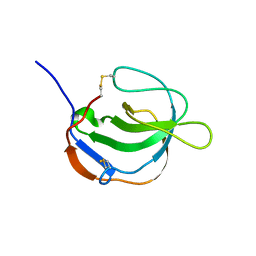 | | STRUCTURE OF MELANOMA INHIBITORY ACTIVITY PROTEIN: A MEMBER OF A NEW FAMILY OF SECRETED PROTEINS | | Descriptor: | MELANOMA DERIVED GROWTH REGULATORY PROTEIN | | Authors: | Lougheed, J.C, Holton, J.M, Alber, T, Bazan, J.F, Handel, T.M. | | Deposit date: | 2001-02-02 | | Release date: | 2001-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structure of melanoma inhibitory activity protein, a member of a recently identified family of secreted proteins.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3CSU
 
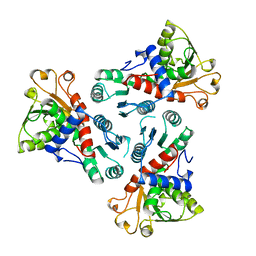 | | CATALYTIC TRIMER OF ESCHERICHIA COLI ASPARTATE TRANSCARBAMOYLASE | | Descriptor: | CALCIUM ION, PROTEIN (ASPARTATE CARBAMOYLTRANSFERASE) | | Authors: | Beernink, P.T, Endrizzi, J.A, Alber, T, Schachman, H.K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-05-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Assessment of the allosteric mechanism of aspartate transcarbamoylase based on the crystalline structure of the unregulated catalytic subunit.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6CCD
 
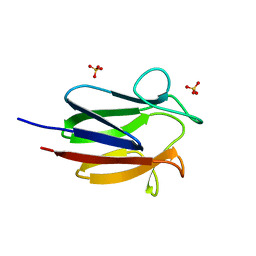 | |
2NT3
 
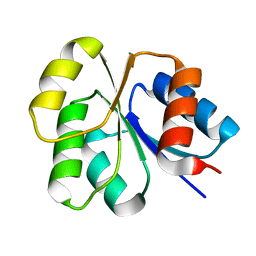 | | Receiver domain from Myxococcus xanthus social motility protein FrzS (Y102A Mutant) | | Descriptor: | Response regulator homolog | | Authors: | Fraser, J.S, Echols, N, Merlie, J.P, Zusman, D.R, Alber, T. | | Deposit date: | 2006-11-06 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
4M6G
 
 | | Structure of the Mycobacterium tuberculosis peptidoglycan amidase Rv3717 in complex with L-Alanine-iso-D-Glutamine reaction product | | Descriptor: | ALANINE, D-alpha-glutamine, Peptidoglycan Amidase Rv3717, ... | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4M6I
 
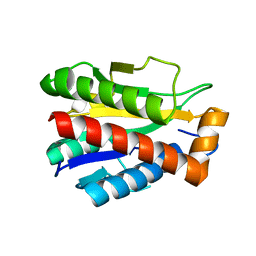 | | Structure of the reduced, Zn-bound form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717, ZINC ION | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
