8JY9
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JYB
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 | | Descriptor: | Butyrophylin 3, SULFATE ION | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
6ONO
 
 | | Complex structure of WhiB1 and region 4 of SigA in C2221 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
7XSI
 
 | |
6ONU
 
 | | Complex structure of WhiB1 and region 4 of SigA in P21 space group. | | Descriptor: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | Authors: | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
4PSX
 
 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
4PSW
 
 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H4 type VIII, Histone acetyltransferase type B catalytic subunit, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
7Y8M
 
 | | Structure of ScIRED-R2-V3 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 2-[2,5-bis(fluoranyl)phenyl]pyrrolidine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, reductase | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
5HDM
 
 | | Crystal structure of Arabidopsis thaliana glutamate-1-semialdehyde-2,1-aminomutase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase 1, chloroplastic, ... | | Authors: | Song, Y, Pu, H, Liu, L. | | Deposit date: | 2016-01-05 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of glutamate-1-semialdehyde-2,1-aminomutase from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3WJ7
 
 | | Crystal structure of gox2253 | | Descriptor: | MERCURY (II) ION, Putative oxidoreductase | | Authors: | Yuan, Y.A, Yin, B. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into substrate and coenzyme preference by SDR family protein Gox2253 from Gluconobater oxydans.
Proteins, 82, 2014
|
|
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
7QUB
 
 | | EV-A71-3Cpro in complex with inhibitor MG78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, Protease 3C, SODIUM ION | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-01-17 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
7QL8
 
 | | SARS-COV2 Main Protease in complex with inhibitor MG78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
7QG5
 
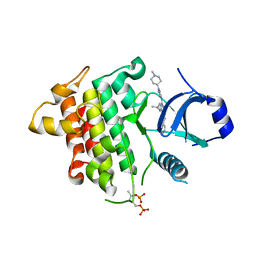 | | IRAK4 in complex with inhibitor | | Descriptor: | 4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]-6-pyrimidin-5-yl-pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
7QG2
 
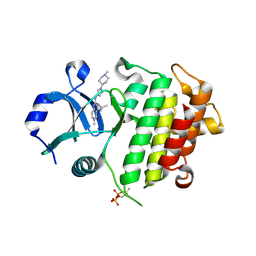 | | IRAK4 in complex with inhibitor | | Descriptor: | 6-methyl-4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.031 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
7QG1
 
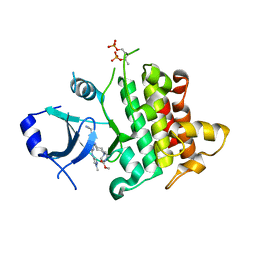 | | IRAK4 in complex with inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, methyl 4-[4-[[6-(cyanomethyl)-2-[(1-methylpyrazol-4-yl)amino]-5-oxidanylidene-pyrido[4,3-d]pyrimidin-4-yl]amino]cyclohexyl]piperazine-1-carboxylate | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
7QG3
 
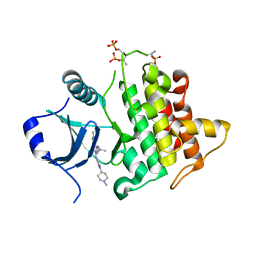 | | IRAK4 in complex with inhibitor | | Descriptor: | 6-[(2~{S})-2-fluoranylpropyl]-4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
4YSJ
 
 | |
4FZW
 
 | | Crystal Structure of the PaaF-PaaG Hydratase-Isomerase Complex from E.coli | | Descriptor: | 1,2-epoxyphenylacetyl-CoA isomerase, 2,3-dehydroadipyl-CoA hydratase, GLYCEROL | | Authors: | Grishin, A.M, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-07-08 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Protein-Protein Interactions in the beta-Oxidation Part of the Phenylacetate
Utilization Pathway. Crystal Structure of the PaaF-PaaG Hydratase-Isomerase Complex
J.Biol.Chem., 287, 2012
|
|
4AQ3
 
 | | HUMAN BCL-2 WITH PHENYLACYLSULFONAMIDE INHIBITOR | | Descriptor: | APOPTOSIS REGULATOR BCL-2, BCL-2-LIKE PROTEIN 1, N,N-dibutyl-4-chloranyl-1-[2-(3,4-dihydro-1H-isoquinolin-2-ylcarbonyl)-4-[(7-iodanylnaphthalen-2-yl)sulfonylcarbamoyl]phenyl]-5-methyl-pyrazole-3-carboxamide | | Authors: | Bertrand, J.A, Fasolini, M, Modugno, M. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a Phenylacylsulfonamide Series of Dual Bcl-2/Bcl-Xl Antagonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4Q9V
 
 | | Crystal structure of TIPE3 | | Descriptor: | CHLORIDE ION, SULFATE ION, Tumor necrosis factor alpha-induced protein 8-like protein 3 | | Authors: | Wu, J, Zhang, X, Chen, Y.H, Shi, Y. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Cancer Cell, 26, 2014
|
|
7YHS
 
 | | Structure of Csy-AcrIF4-dsDNA | | Descriptor: | AcrIF4, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Feng, Y, Zhang, L.X. | | Deposit date: | 2022-07-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Anti-CRISPR protein AcrIF4 inhibits the type I-F CRISPR-Cas surveillance complex by blocking nuclease recruitment and DNA cleavage.
J.Biol.Chem., 298, 2022
|
|
4WN9
 
 | | Structure of the Nitrogenase MoFe Protein from Clostridium pasteurianum Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
6E66
 
 | |
6M3M
 
 | |
