1QMF
 
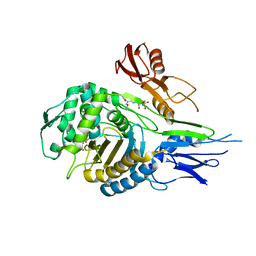 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
1GRG
 
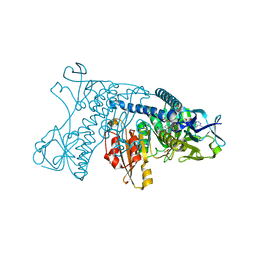 | |
1GRE
 
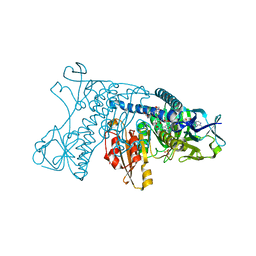 | |
2H92
 
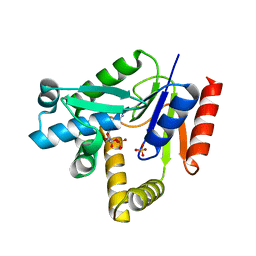 | |
6RS2
 
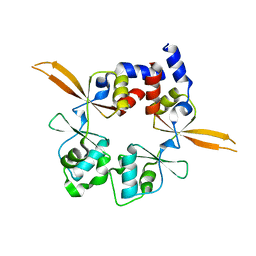 | | Structure of the Bateman module of human CNNM4. | | Descriptor: | Metal transporter CNNM4 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Gomez-Garcia, I, Oyenarte, I, Gimenez, P, Ereno-Orbea, J, Diercks, T, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2019-05-21 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.694 Å) | | Cite: | Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci, 20, 2019
|
|
7KZ9
 
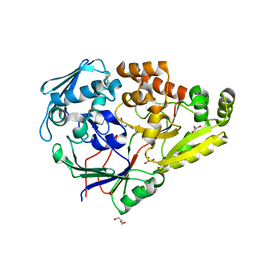 | | Crystal structure of Pseudomonas sp. PDC86 substrate-binding protein Aapf in complex with a signaling molecule HEHEAA | | Descriptor: | GLYCEROL, N,N~2~-bis(2-hydroxyethyl)glycinamide, Peptide/nickel transport system substrate-binding protein AapF, ... | | Authors: | Luo, S, Dadhwal, P, Tong, L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a bacterial Pip system plant effector recognition protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KZ8
 
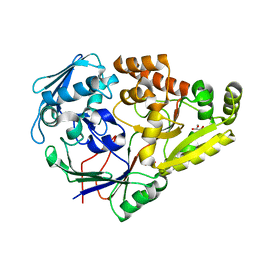 | |
4RV7
 
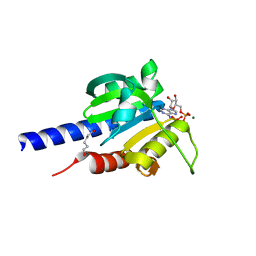 | | Characterization of an essential diadenylate cyclase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Diadenylate cyclase, HEXANE-1,6-DIOL, ... | | Authors: | Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2014-11-25 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Biochemical Analysis of the Essential Diadenylate Cyclase CdaA from Listeria monocytogenes.
J.Biol.Chem., 290, 2015
|
|
6VTN
 
 | |
6VTY
 
 | |
9WGA
 
 | |
5K7K
 
 | |
6UX5
 
 | |
6XRJ
 
 | |
6G52
 
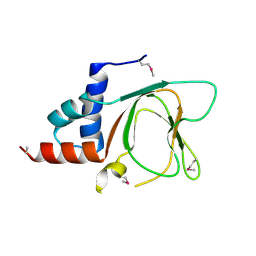 | | CRYSTAL STRUCTURE OF THE CNMP BINDING DOMAIN OF THE MAGNESIUM TRANSPORTER CNNM4 | | Descriptor: | Metal transporter CNNM4 | | Authors: | Gimenez, P, Oyenarte, I, Hardy, S, Zubillaga, M, Merino, N, Blanco, F.J, Siliqi, D, Tremblay, M, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2018-03-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.691 Å) | | Cite: | Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci, 20, 2019
|
|
6DB3
 
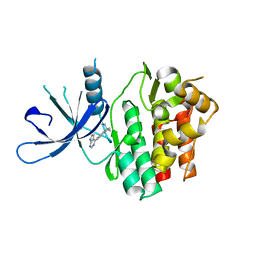 | | JAK3 with Cyanamide CP23 | | Descriptor: | Tyrosine-protein kinase JAK3, [(1S)-1-methyl-6-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]cyanamide | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-02 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Identification of Cyanamide-Based Janus Kinase 3 (JAK3) Covalent Inhibitors.
J. Med. Chem., 61, 2018
|
|
6DUD
 
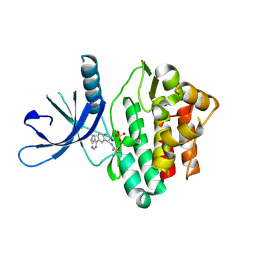 | | JAK3 with cyanamide CP12 | | Descriptor: | N-[(1S)-6-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]imidoformamide, SULFATE ION, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-06-20 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Identification of Cyanamide-Based Janus Kinase 3 (JAK3) Covalent Inhibitors.
J. Med. Chem., 61, 2018
|
|
6DB4
 
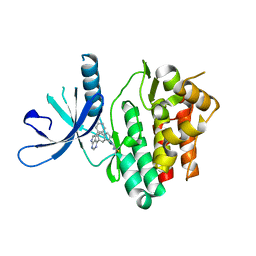 | | JAK3 with Cyanamide CP34 | | Descriptor: | N-[(1S)-6-(5-phenyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]imidoformamide, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-02 | | Release date: | 2018-11-28 | | Last modified: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Identification of Cyanamide-Based Janus Kinase 3 (JAK3) Covalent Inhibitors.
J. Med. Chem., 61, 2018
|
|
6DA4
 
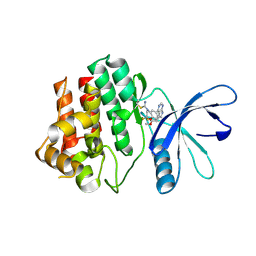 | | JAK3 with Cyanamide CP10 | | Descriptor: | (Z)-1-{2,2-difluoro-6-[5-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-2,3-dihydro-4H-1,4-benzoxazin-4-yl}methanimine, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of Cyanamide-Based Janus Kinase 3 (JAK3) Covalent Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CKF
 
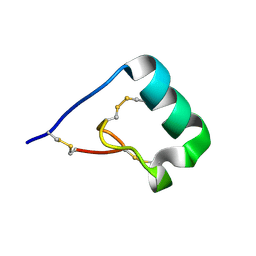 | | Structure of a new ShKT peptide from the sea anemone Oulactis sp: OspTx2a-p2 | | Descriptor: | OspTx2a-p2 | | Authors: | Sunanda, P, Krishnarjuna, B, Norton, R.S. | | Deposit date: | 2018-02-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Identification, chemical synthesis, structure, and function of a new KV1 channel blocking peptide from Oulactis sp.
Peptide Science, 110, 2018
|
|
6CKD
 
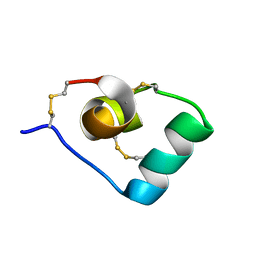 | | Structure of a new ShKT peptide from the sea anemone Oulactis sp: OspTx2a-p1 | | Descriptor: | OspTx2a-p1 | | Authors: | Sunanda, P, Krishnarjuna, B, Norton, R.S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification, chemical synthesis, structure, and function of a new KV1 channel blocking peptide from Oulactis sp.
Peptide Science, 110, 2018
|
|
6BUC
 
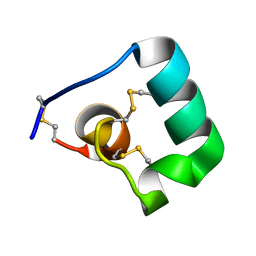 | |
6UIP
 
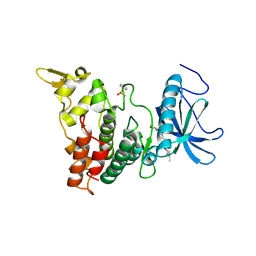 | | DYRK1A Kinase Domain in Complex with a 6-azaindole Derivative, GNF2133. | | Descriptor: | 4-ethyl-N-{4-[1-(oxan-4-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]pyridin-2-yl}piperazine-1-carboxamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133.
J.Med.Chem., 63, 2020
|
|
6MRK
 
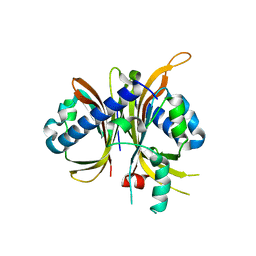 | |
6OPF
 
 | |
