1GHS
 
 | |
1GHR
 
 | |
6FWB
 
 | | Crystal structure of Mat2A at 1.79 Angstron resolution | | Descriptor: | GLYCEROL, S-adenosylmethionine synthase isoform type-2, SODIUM ION, ... | | Authors: | Zhou, A, Wei, Z, Bai, J, Wang, H. | | Deposit date: | 2018-03-06 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of a natural inhibitor of methionine adenosyltransferase 2A regulating one-carbon metabolism in keratinocytes.
Ebiomedicine, 39, 2019
|
|
5DH3
 
 | | Crystal structure of MST2 in complex with XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2015-08-29 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration
Sci Transl Med, 8, 2016
|
|
4QR3
 
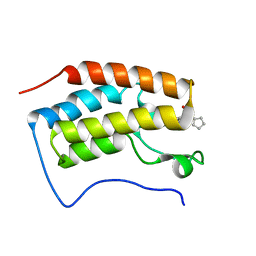 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-cyclopentyl-3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR5
 
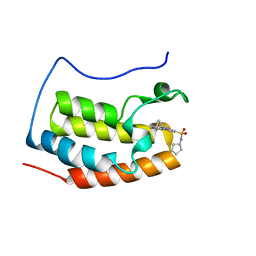 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
9JG6
 
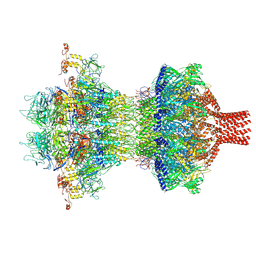 | | The tail-complex structure of phage P22 | | Descriptor: | Endorhamnosidase, P22 tail accessory factor, Phage stabilisation protein, ... | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-09-06 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9JGA
 
 | | P22 procapsid portal | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-09-06 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYV
 
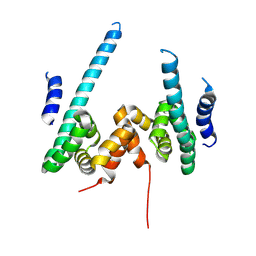 | | The scaffold dimer of phage P22 | | Descriptor: | Scaffolding protein | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-12-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYW
 
 | | The scaffold C-loop of phage P22 | | Descriptor: | Portal protein, Scaffolding protein | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-12-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYY
 
 | | The scaffold trimer of phage P22 | | Descriptor: | Scaffolding protein | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-12-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
9KYX
 
 | | The scaffold tetramer of phage P22 | | Descriptor: | Scaffolding protein | | Authors: | Liu, H.R, Xiao, H. | | Deposit date: | 2024-12-09 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of the scaffolding protein and portal within the bacteriophage P22 procapsid provides insights into the self-assembly process.
Plos Biol., 23, 2025
|
|
6LZH
 
 | | Crystal structure of Alpha/beta hydrolase GrgF from Penicillium sp. sh18 | | Descriptor: | GrgF, SODIUM ION | | Authors: | Wang, H, Yu, J, Wang, W.G, Matsuda, Y, Yao, M. | | Deposit date: | 2020-02-19 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for the Biosynthesis of an Unusual Chain-Fused Polyketide, Gregatin A.
J.Am.Chem.Soc., 142, 2020
|
|
4FZ3
 
 | | Crystal structure of SIRT3 in complex with acetyl p53 peptide coupled with 4-amino-7-methylcoumarin | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ZINC ION, ... | | Authors: | Liu, D, Wu, J, Zhang, D, Chen, K, Jiang, H, Liu, H. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanism Study of SIRT1 Activators that Promote the Deacetylation of Fluorophore-Labeled Substrate
J.Med.Chem., 56, 2013
|
|
5HZG
 
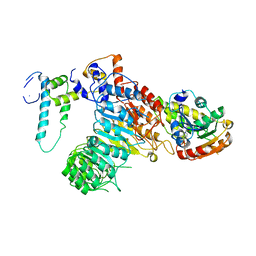 | | The crystal structure of the strigolactone-induced AtD14-D3-ASK1 complex | | Descriptor: | (2Z)-2-methylbut-2-ene-1,4-diol, F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A, ... | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
5HYW
 
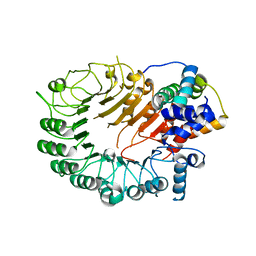 | | The crystal structure of the D3-ASK1 complex | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
2WH6
 
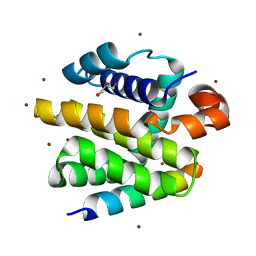 | | Crystal structure of anti-apoptotic BHRF1 in complex with the Bim BH3 domain | | Descriptor: | BCL-2-LIKE PROTEIN 11, BROMIDE ION, EARLY ANTIGEN PROTEIN R, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2009-05-01 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1.
PLoS Pathog., 6, 2010
|
|
2V6Q
 
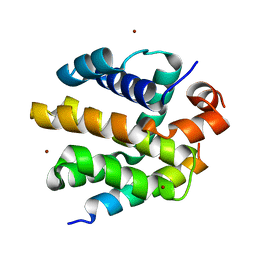 | |
2XPX
 
 | | Crystal structure of BHRF1:Bak BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BHRF1, BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus Bhrf1.
Plos Pathog., 6, 2010
|
|
3UBU
 
 | | Crystal structure of agkisacucetin, a GpIb-binding snaclec (snake C-type lectin) that inhibits platelet | | Descriptor: | Agglucetin subunit alpha-1, Agglucetin subunit beta-2, GLYCEROL, ... | | Authors: | Gao, Y, Ge, H, Chen, H, Li, H, Liu, Y, Niu, L, Teng, M. | | Deposit date: | 2011-10-25 | | Release date: | 2012-04-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of agkisacucetin, a Gpib-binding snake C-type lectin that inhibits platelet adhesion and aggregation.
Proteins, 2012
|
|
4R8T
 
 | | Structure of JEV protease | | Descriptor: | CHLORIDE ION, NS3, Serine protease subunit NS2B | | Authors: | Nair, D.T, Weinert, T, Wang, M, Olieric, V. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4R8U
 
 | | S-SAD structure of DINB-DNA Complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA, DNA polymerase IV, ... | | Authors: | Kottur, J, Nair, D.T, Weinert, T, Oligeric, V, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination
Nat.Methods, 12, 2015
|
|
7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | Descriptor: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2L
 
 | | Crystal structure of nanobody 3-2A2-4 with SARS-CoV-2 RBD | | Descriptor: | Nanobody 3-2A2-4, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
