4QVX
 
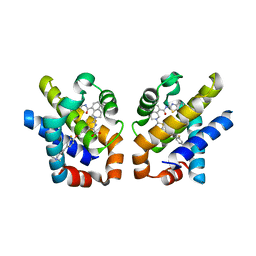 | |
5YMU
 
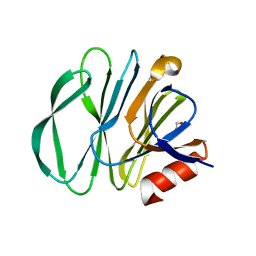 | |
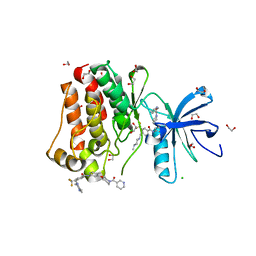
5HU9
 
 | | Crystal structure of ABL1 in complex with CHMFL-074 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-methylpiperazin-1-yl)methyl]-N-(4-methyl-3-{[1-(pyridin-3-ylcarbonyl)piperidin-4-yl]oxy}phenyl)-3-(trifluoromethyl)benzamide, CHLORIDE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2016-01-27 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Discovery and characterization of a novel potent type II native and mutant BCR-ABL inhibitor (CHMFL-074) for Chronic Myeloid Leukemia (CML)
Oncotarget, 7, 2016
|
|
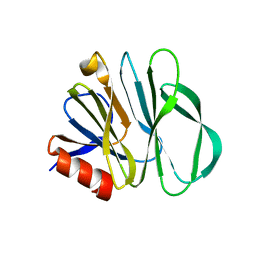
5YMT
 
 | |
5VAG
 
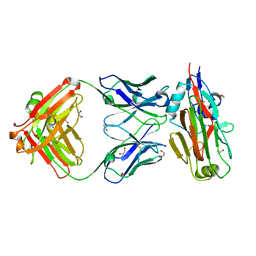 | | Crystal structure of H7-specific antibody m826 in complex with the HA1 domain of hemagglutinin from H7N9 influenza virus | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antibody m826, Hemagglutinin, ... | | Authors: | Song, H, Ying, T, Ji, X. | | Deposit date: | 2017-03-26 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Potent Germline-like Human Monoclonal Antibody Targets a pH-Sensitive Epitope on H7N9 Influenza Hemagglutinin.
Cell Host Microbe, 22, 2017
|
|
3KME
 
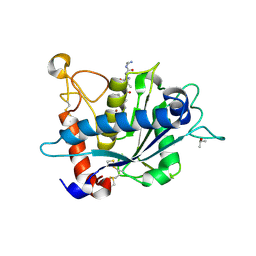 | | Crystal structure of catalytic domain of TACE with phenyl-pyrrolidinyl-tartrate inhibitor | | Descriptor: | (2R,3R)-2,3-dihydroxy-4-oxo-4-[(2R)-2-phenylpyrrolidin-1-yl]-N-(thiophen-2-ylmethyl)butanamide, ISOPROPYL ALCOHOL, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, ... | | Authors: | Orth, P. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The discovery of novel tartrate-based TNF-alpha converting enzyme (TACE) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2009
|
|
3L59
 
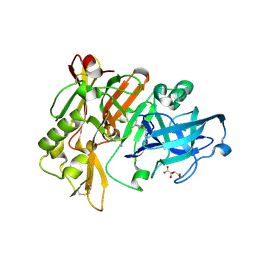 | | Structure of BACE Bound to SCH710413 | | Descriptor: | (2Z)-3-(3-chlorobenzyl)-2-imino-5,5-dimethylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2023-01-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
6XZ8
 
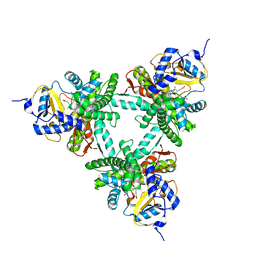 | | Structure of aldosterone synthase (CYP11B2) in complex with N-[(1R)-1-[5-(6-chloro-1,1-dimethyl-3-oxo-isoindolin-2-yl)-3-pyridyl]ethyl]methanesulfonamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, HEME C, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6XZ9
 
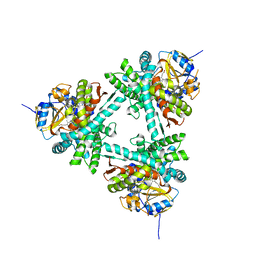 | | Structure of aldosterone synthase (CYP11B2) in complex with 5-chloro-3,3-dimethyl-2-[5-[1-(1-methylpyrazole-4-carbonyl)azetidin-3-yl]oxy-3-pyridyl]isoindolin-1-one | | Descriptor: | 5-chloranyl-3,3-dimethyl-2-[5-[1-(1-methylpyrazol-4-yl)carbonylazetidin-3-yl]oxypyridin-3-yl]isoindol-1-one, Cytochrome P450 11B2, mitochondrial, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
3L58
 
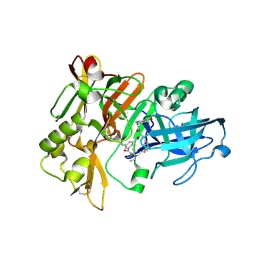 | | Structure of BACE Bound to SCH589432 | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5E
 
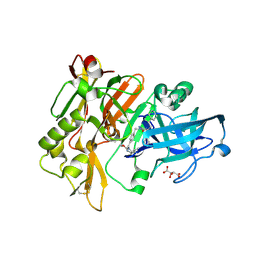 | | Structure of BACE Bound to SCH736062 | | Descriptor: | (4S)-1-(4-{[(2Z,4R)-4-(2-cyclohexylethyl)-4-(cyclohexylmethyl)-2-imino-5-oxoimidazolidin-1-yl]methyl}benzyl)-4-propylimidazolidin-2-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
6E9L
 
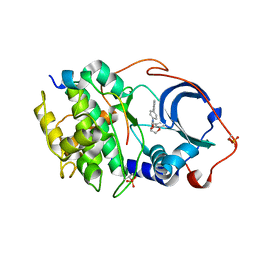 | |
6E9W
 
 | | Crystal structure of Rock1 with a pyridinylbenzamide based inhibitor | | Descriptor: | N-[(2,3-dihydro-1,4-benzodioxin-5-yl)methyl]-4-(pyridin-4-yl)benzamide, Rho-associated protein kinase 1, SULFATE ION | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification of Selective Dual ROCK1 and ROCK2 Inhibitors Using Structure Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6ED6
 
 | |
3L5D
 
 | | Structure of BACE Bound to SCH723873 | | Descriptor: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5C
 
 | | Structure of BACE Bound to SCH723871 | | Descriptor: | 1-(4-cyanophenyl)-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5F
 
 | | Structure of BACE Bound to SCH736201 | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-(cyclohexylmethyl)-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
6IEG
 
 | | Crystal structure of human MTR4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Exosome RNA helicase MTR4, MAGNESIUM ION | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
2R2J
 
 | | crystal structure of human ERp44 | | Descriptor: | FORMIC ACID, SUCCINIC ACID, Thioredoxin domain-containing protein 4 | | Authors: | Wang, L.K, Li, S.J, Sun, F, Wang, C.C. | | Deposit date: | 2007-08-25 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail.
Embo Rep., 2008
|
|
6IEH
 
 | | Crystal structures of the hMTR4-NRDE2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
3L5B
 
 | | Structure of BACE Bound to SCH713601 | | Descriptor: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3KMC
 
 | | Crystal structure of catalytic domain of TACE with tartrate-based inhibitor | | Descriptor: | (2R,3R)-4-[4-(2-chlorophenyl)piperazin-1-yl]-2,3-dihydroxy-4-oxo-N-(2-thiophen-2-ylethyl)butanamide, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, TNF-alpha-converting enzyme, ... | | Authors: | Orth, P. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The discovery of novel tartrate-based TNF-alpha converting enzyme (TACE) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2009
|
|
6E99
 
 | |
8YQQ
 
 | | Structure of HKU1B RBD with TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, Transmembrane protease serine 2 | | Authors: | Gao, X, Cui, S, Ding, W, Zhu, K, Shang, K, Zhu, H. | | Deposit date: | 2024-03-19 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis for the interaction between human coronavirus HKU1 spike receptor binding domain and its receptor TMPRSS2.
Cell Discov, 10, 2024
|
|
