3C6B
 
 | |
3KK1
 
 | |
3KK3
 
 | | HIV-1 reverse transcriptase-DNA complex with GS-9148 terminated primer | | Descriptor: | 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*C*(URT))-3', 5'-D(*AP*TP*GP*GP*TP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2009-11-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex.
J.Mol.Biol., 397, 2010
|
|
3KJV
 
 | | HIV-1 reverse transcriptase in complex with DNA | | Descriptor: | 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DOC))-3', 5'-D(*AP*TP*GP*GP*TP*GP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2009-11-03 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex.
J.Mol.Biol., 397, 2010
|
|
3KK2
 
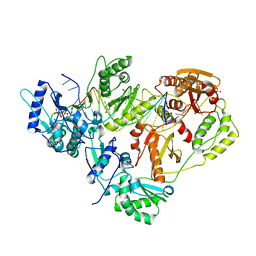 | | HIV-1 reverse transcriptase-DNA complex with dATP bound in the nucleotide binding site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*A*TP*GP*GP*TP*GP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', 5'-D(*AP*CP*A*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DOC))-3', ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2009-11-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex.
J.Mol.Biol., 397, 2010
|
|
3H7V
 
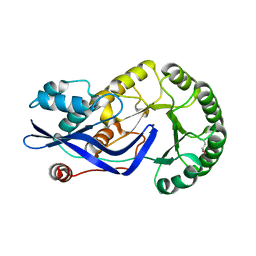 | | CRYSTAL STRUCTURE OF O-SUCCINYLBENZOATE SYNTHASE FROM THERMOSYNECHOCOCCUS ELONGATUS BP-1 complexed with MG in the active site | | Descriptor: | MAGNESIUM ION, O-SUCCINYLBENZOATE SYNTHASE | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3H70
 
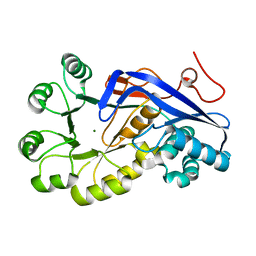 | | Crystal structure of o-succinylbenzoic acid synthetase from staphylococcus aureus Complexed with mg in the active site | | Descriptor: | MAGNESIUM ION, O-succinylbenzoic acid (OSB) synthetase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1F9I
 
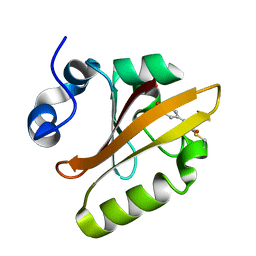 | | CRYSTAL STRUCTURE OF THE PHOTOACTIVE YELLOW PROTEIN MUTANT Y42F | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Brudler, R, Meyer, T.E, Genick, U.K, Tollin, G, Getzoff, E.D. | | Deposit date: | 2000-07-10 | | Release date: | 2000-07-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein.
Biochemistry, 39, 2000
|
|
1F98
 
 | | CRYSTAL STRUCTURE OF THE PHOTOACTIVE YELLOW PROTEIN MUTANT T50V | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Brudler, R, Meyer, T.E, Genick, U.K, Tollin, G, Getzoff, E.D. | | Deposit date: | 2000-07-07 | | Release date: | 2000-07-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein.
Biochemistry, 39, 2000
|
|
3CAW
 
 | | Crystal structure of o-succinylbenzoate synthase from Bdellovibrio bacteriovorus liganded with Mg | | Descriptor: | MAGNESIUM ION, o-succinylbenzoate synthase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1DIW
 
 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH GALACTOSE | | Descriptor: | TETANUS TOXIN HC, beta-D-galactopyranose | | Authors: | Emsley, P, Fotinou, C, Black, I, Fairweather, N.F, Charles, I.G, Watts, C, Hewitt, E, Isaacs, N.W. | | Deposit date: | 1999-11-30 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the H(C) fragment of tetanus toxin with carbohydrate subunit complexes provide insight into ganglioside binding.
J.Biol.Chem., 275, 2000
|
|
1DFQ
 
 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH SIALIC ACID | | Descriptor: | N-acetyl-beta-neuraminic acid, TETANUS TOXIN HC | | Authors: | Emsley, P, Fotinou, C, Black, I, Fairweather, N.F, Charles, I.G, Watts, C, Hewitt, E, Isaacs, N.W. | | Deposit date: | 1999-11-20 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structures of the H(C) fragment of tetanus toxin with carbohydrate subunit complexes provide insight into ganglioside binding.
J.Biol.Chem., 275, 2000
|
|
1WUE
 
 | | Crystal structure of protein GI:29375081, unknown member of enolase superfamily from enterococcus faecalis V583 | | Descriptor: | mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-05 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1WUF
 
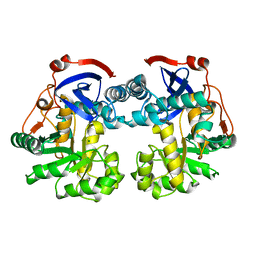 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1D0H
 
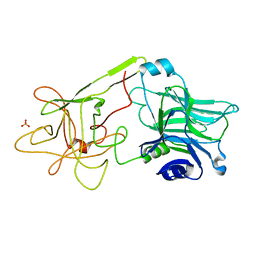 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH N-ACETYL-GALACTOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, PROTEIN (TETANUS TOXIN HC), SULFATE ION | | Authors: | Emsley, P, Fotinou, C, Black, I, Fairweather, N.F, Charles, I.G, Watts, C, Hewitt, E, Isaacs, N.W. | | Deposit date: | 1999-09-10 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structures of the H(C) fragment of tetanus toxin with carbohydrate subunit complexes provide insight into ganglioside binding.
J.Biol.Chem., 275, 2000
|
|
1DLL
 
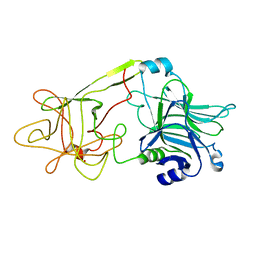 | | The HC fragement of tetanus toxin complexed with lactose | | Descriptor: | GLYCEROL, TETANUS TOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Emsley, P, Fotinou, C, Black, I, Fairweather, N.F, Charles, I.G, Watts, C, Hewitt, E, Isaacs, N.W. | | Deposit date: | 1999-12-10 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of the H(C) fragment of tetanus toxin with carbohydrate subunit complexes provide insight into ganglioside binding.
J.Biol.Chem., 275, 2000
|
|
2I4X
 
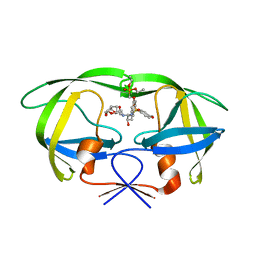 | | HIV-1 Protease I84V, L90M with GS-8374 | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
1GSU
 
 | | AN AVIAN CLASS-MU GLUTATHIONE S-TRANSFERASE, CGSTM1-1 AT 1.94 ANGSTROM RESOLUTION | | Descriptor: | CLASS-MU GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Sun, Y.-J, Kuan, C, Tam, M.F, Hsiao, C.-D. | | Deposit date: | 1997-09-02 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The three-dimensional structure of an avian class-mu glutathione S-transferase, cGSTM1-1 at 1.94 A resolution.
J.Mol.Biol., 278, 1998
|
|
1YKC
 
 | |
1FV2
 
 | | The Hc fragment of tetanus toxin complexed with an analogue of its ganglioside receptor GT1B | | Descriptor: | ETHYL-TRIMETHYL-SILANE, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Fotinou, C, Emsley, P, Black, I, Ando, H, Ishida, H, Kiso, M, Sinha, K.A, Fairweather, N.F, Isaacs, N.W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of tetanus toxin Hc fragment complexed with a synthetic GT1b analogue suggests cross-linking between ganglioside receptors and the toxin.
J.Biol.Chem., 276, 2001
|
|
1XW5
 
 | |
1FV3
 
 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH AN ANALOGUE OF ITS GANGLIOSIDE RECEPTOR GT1B | | Descriptor: | ETHYL-TRIMETHYL-SILANE, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Fotinou, C, Emsley, P, Black, I, Ando, H, Ishida, H, Kiso, M, Sinha, K.A, Fairweather, N.F, Isaacs, N.W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of tetanus toxin Hc fragment complexed with a synthetic GT1b analogue suggests cross-linking between ganglioside receptors and the toxin.
J.Biol.Chem., 276, 2001
|
|
2AB6
 
 | |
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A5U
 
 | | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex | | Descriptor: | DNA (31-MER), Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Manjunath, G.P, Sekar, K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2009-08-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Promiscuity and specificity in DNA binding to SSB: Insights from the structure of the Mycobacterium smegmatis SSB-ssDNA complex.
To be Published, 2009
|
|
