6YGS
 
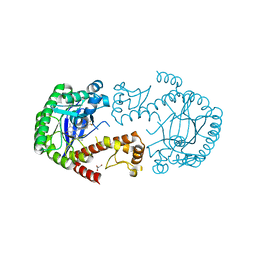 | | tRNA-guanine Transglycosylase (TGT) in co-crystallized complex with 6-amino-2-(methylamino)-4-(4-(trifluoromethyl)phenethyl)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-(methylamino)-4-(4-(trifluoromethyl)phenethyl)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-03-27 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Co-crystallization and 19F NMR reveal dimer disturbing inhibitors and conformational changes at dimer contacts
To Be Published
|
|
6YHD
 
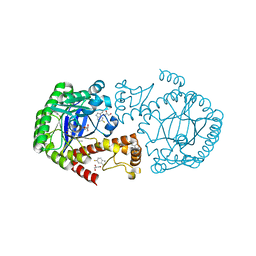 | | tRNA-guanine Transglycosylase (TGT) labeled with 5-fluorotryptophan in co-crystallized complex with 6-amino-4-(2-((2R,3S,4R,5R)-3,4-dihydroxy-5-methoxytetrahydrofuran-2-yl)ethyl)-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Nguyen, A, Heine, A, Klebe, G. | | Deposit date: | 2020-03-28 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Co-crystallization, nanoESI-MS and 19F NMR reveal dimer disturbing inhibitors and conformational changes at dimer contacts
To Be Published
|
|
6Y03
 
 | |
3M04
 
 | | Carbonic Anhydrase II in complex with novel sulfonamide inhibitor | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, ZINC ION, ... | | Authors: | Schulze Wischeler, J, Heine, A, Klebe, G, Sandner, N.U. | | Deposit date: | 2010-03-02 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural investigation and inhibitor studies on Carbonic Anhydrase II
To be Published
|
|
3M1K
 
 | | Carbonic Anhydrase in complex with fragment | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 1-hydroxy-2-sulfanylpyridinium, Carbonic anhydrase 2, ... | | Authors: | Schulze Wischeler, J, Heine, A, Klebe, G. | | Deposit date: | 2010-03-05 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Bidentate Zinc chelators for alpha-carbonic anhydrases that produce a trigonal bipyramidal coordination geometry.
Chemmedchem, 5, 2010
|
|
3M2Y
 
 | |
3M1Q
 
 | |
3M2Z
 
 | |
3M3D
 
 | | Crystal structure of Acetylcholinesterase in complex with Xenon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Behnen, J, Brumshtein, B, Toker, L, Silman, I, Sussman, J.L, Klebe, G, Heine, A. | | Deposit date: | 2010-03-09 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Old acquantance rediscover; use of xenon/protein complexes as a generic tool for SAD phasing of inhouse data
To be Published
|
|
3M4H
 
 | | Human Aldose Reductase mutant T113V complexed with IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, Aldose reductase, BROMIDE ION, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-11 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3M5S
 
 | |
3LZ5
 
 | | Human aldose reductase mutant T113V complexed with IDD594 | | Descriptor: | Aldose Reductase, CITRIC ACID, IDD594, ... | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-01 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Tracing the detail: how mutations affect binding modes and thermodynamic signatures of closely related aldose reductase inhibitors
J.Mol.Biol., 406, 2011
|
|
3LZY
 
 | |
3M14
 
 | | Carbonic Anhydrase II in complex with novel sulfonamide inhibitor | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase 2, N-[(2Z)-1,3-thiazolidin-2-ylidene]sulfamide, ... | | Authors: | Schulze Wischeler, J, Heine, A, Klebe, G, Sandner, N.U. | | Deposit date: | 2010-03-04 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural investigation and inhibitor studies on Carbonic Anhydrase II
To be Published
|
|
3PRS
 
 | | Endothiapepsin in complex with ritonavir | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-30 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
3PCZ
 
 | | Endothiapepsin in complex with benzamidine | | Descriptor: | BENZAMIDINE, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental and computational active site mapping as a starting point to fragment-based lead discovery.
Chemmedchem, 7, 2012
|
|
2Z7K
 
 | | tRNA-Guanine transglycosylase (TGT) in complex with 2-Amino-lin-Benzoguanine | | Descriptor: | 2,6-diamino-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2007-08-24 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure analysis and in silico pKa calculations suggest strong pKa shifts of ligands as driving force for high-affinity binding to TGT
Chembiochem, 10, 2009
|
|
6QL0
 
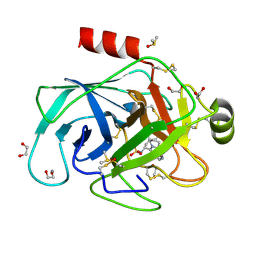 | |
2ZDM
 
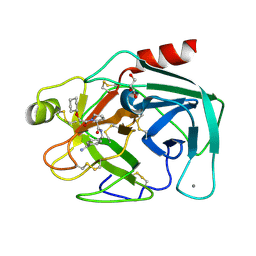 | | Exploring trypsin S3 pocket | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexyloxy)ethanoyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Baum, B, Brandt, T, Heine, A, Klebe, G. | | Deposit date: | 2007-11-26 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties
J.Mol.Biol., 405, 2011
|
|
2ZGX
 
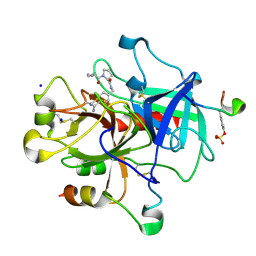 | | Thrombin Inhibition | | Descriptor: | 1-[(2R)-2-aminobutanoyl]-N-(4-carbamimidoylbenzyl)-L-prolinamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-28 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin
J.Mol.Biol., 391, 2009
|
|
2PD5
 
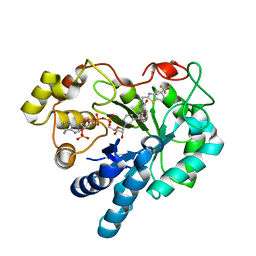 | | Human aldose reductase mutant V47I complexed with zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2ZFR
 
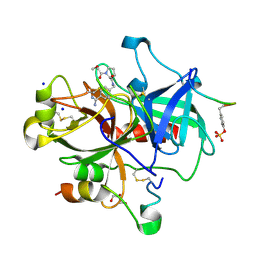 | | Exploring thrombin S3 pocket | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexyloxy)ethanoyl)pyrrolidine-2-carboxamide, GLYCEROL, Hirudin, ... | | Authors: | Brandt, T, Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-10 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploring thrombin S3 pocket
To be Published
|
|
2PDC
 
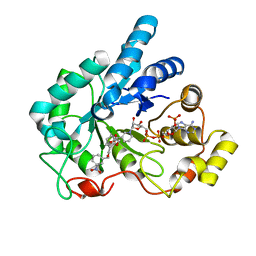 | | Human aldose reductase mutant F121P complexed with IDD393. | | Descriptor: | (5-CHLORO-2-{[(3-NITROBENZYL)AMINO]CARBONYL}PHENOXY)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDK
 
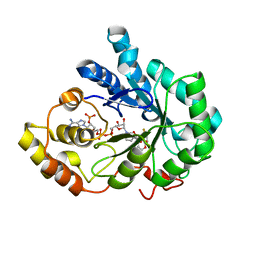 | | Human aldose reductase mutant L301M complexed with sorbinil. | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SORBINIL | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-04-01 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2PDG
 
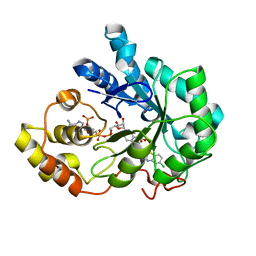 | | Human aldose reductase with uracil-type inhibitor at 1.42A. | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {3-[(5-CHLORO-1,3-BENZOTHIAZOL-2-YL)METHYL]-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL}ACETIC ACID | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
