6PWR
 
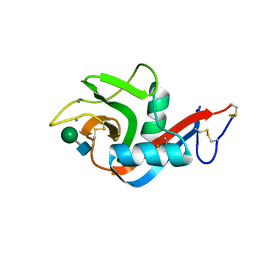 | |
9DLW
 
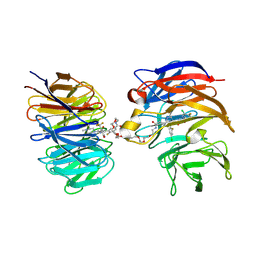 | | Crystal structure of the ternary complex of DCAF1 and WDR5 with PROTAC, OICR-41114 | | Descriptor: | DDB1- and CUL4-associated factor 1, N-{(1P)-5'-({(32E)-33-[(3P)-4-{[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]carbamoyl}-3-(4-chloro-2-fluorophenyl)-1H-pyrrol-2-yl]-31-oxo-3,6,9,12,15,18,21,24,27-nonaoxa-30-azatritriacont-32-en-1-yl}carbamoyl)-2'-fluoro-4-[(3R,5S)-3,4,5-trimethylpiperazin-1-yl][1,1'-biphenyl]-3-yl}-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Mamai, A, Wilson, B.J, Hoffer, L, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-09-11 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
8QWX
 
 | | Ligninolytic manganese peroxidase Ape-MnP1 from Agaricales mushrooms | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2023-10-20 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-function characterization of two enzymes from novel subfamilies of manganese peroxidases secreted by the lignocellulose-degrading Agaricales fungi Agrocybe pediades and Cyathus striatus.
Biotechnol Biofuels Bioprod, 17, 2024
|
|
7BZL
 
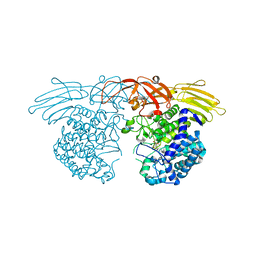 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-04-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8QX0
 
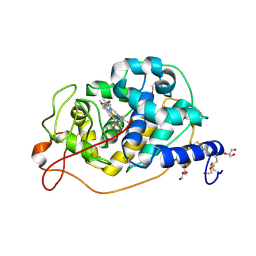 | |
8QWT
 
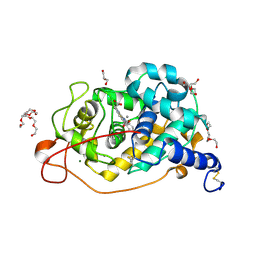 | |
8R1M
 
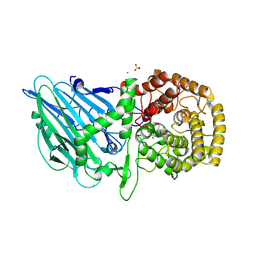 | | Structure of TxGH116 with covalently bound N-azido-octyl aziridine | | Descriptor: | (1R,2R,3R,4S)-4-[8-(azanyldiazenyl)octylamino]-3-(hydroxymethyl)cyclopentane-1,2-diol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Offen, W.A, Davies, G.J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective labelling of GBA2 in cells with fluorescent beta-d-arabinofuranosyl cyclitol aziridines.
Chem Sci, 15, 2024
|
|
6R1N
 
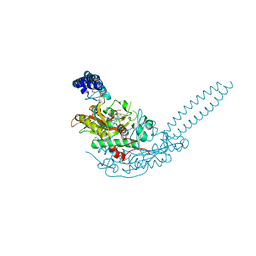 | | Crystal structure of S. aureus seryl-tRNA synthetase complexed to seryl sulfamoyl adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
6R1O
 
 | | Crystal structure of E. coli seryl-tRNA synthetase complexed to a seryl sulfamoyl adenosine derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
6R1M
 
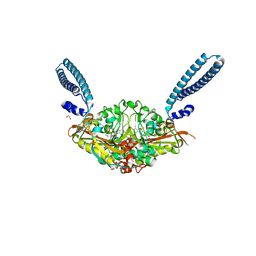 | | Crystal structure of E. coli seryl-tRNA synthetase complexed to seryl sulfamoyl adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, PHOSPHATE ION, ... | | Authors: | Salimraj, R, Cain, R, Roper, D.I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Enhancement of Selectivity of Chemical Probe Inhibitors Targeting Bacterial Seryl-tRNA Synthetase.
J.Med.Chem., 62, 2019
|
|
9DKO
 
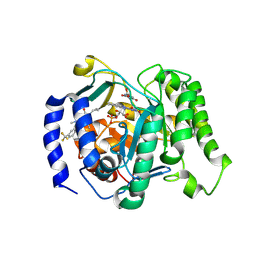 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1010 (6-cyclopropyl-2,4-dimethyl-3-(4-(trifluoromethyl)benzyl)-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, 6-cyclopropyl-2,4-dimethyl-3-{[4-(trifluoromethyl)phenyl]methyl}-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DKY
 
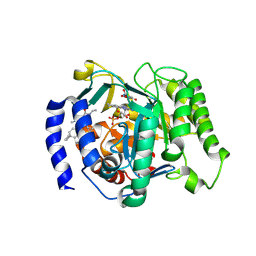 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1174 (3-((7-azaspiro[3.5]nonan-7-yl)methyl)-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | Descriptor: | 3-[(7-azaspiro[3.5]nonan-7-yl)methyl]-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-10 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DLK
 
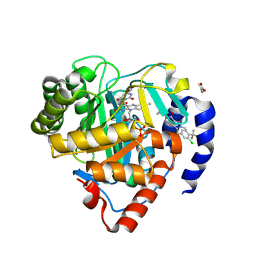 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1398 ((S)-3-(amino(3-chloro-4-(trifluoromethyl)phenyl)(cyclopropyl)methyl)-6-cyclopropyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one) | | Descriptor: | 3-[(R)-amino[3-chloro-4-(trifluoromethyl)phenyl](cyclopropyl)methyl]-6-cyclopropyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, ACETIC ACID, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-11 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DLY
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1211 ((R)-3-(amino(6-(trifluoromethyl)pyridin-3-yl)methyl)-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | Descriptor: | 3-{(R)-amino[6-(trifluoromethyl)pyridin-3-yl]methyl}-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchic, D, Phillips, M. | | Deposit date: | 2024-09-11 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DI6
 
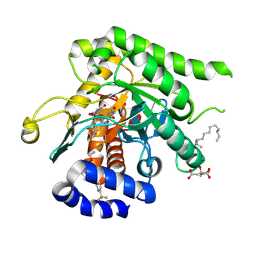 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM679 (ethyl 1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxylate) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, CITRIC ACID, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DKQ
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1153 (4,6-dicyclopropyl-3-(3-fluoro-4-(trifluoromethyl)benzyl)-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one) | | Descriptor: | 4,6-dicyclopropyl-3-{[3-fluoro-4-(trifluoromethyl)phenyl]methyl}-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchic, D, Phillips, M. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
3GLM
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with crotonyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
5UR6
 
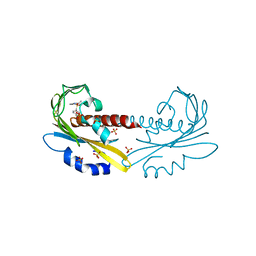 | | PYR1 bound to the rationally designed agonist cyanabactin | | Descriptor: | Abscisic acid receptor PYR1, N-(4-cyano-3-cyclopropylphenyl)-1-(4-methylphenyl)methanesulfonamide, SULFATE ION | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2017-02-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
3GF7
 
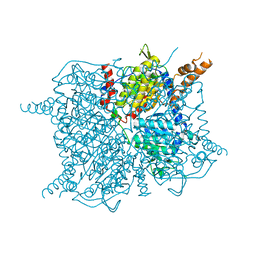 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum apoprotein | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, SULFATE ION | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
9B2M
 
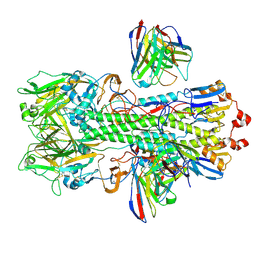 | | Hemagglutinin H1 New Caledonia 1999 in complex with monoclonal antibody Fab 43_S0008 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain monoclonal antibody Fab 3_S0008, Hemagglutinin HA1 chain, ... | | Authors: | Torrents de la Pena, A, de Paiva Froes Rocha, R, Ward, A.B, Ataca, S, Kanekiyo, M. | | Deposit date: | 2024-03-15 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Modulating the immunodominance hierarchy of immunoglobulin germline-encoded structural motifs targeting the influenza hemagglutinin stem.
Cell Rep, 43, 2024
|
|
3GF3
 
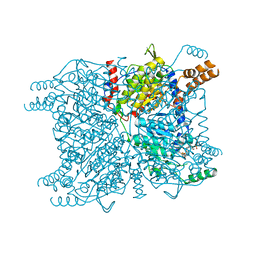 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaconyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GMA
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaryl-CoA | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, glutaryl-coenzyme A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
8UT7
 
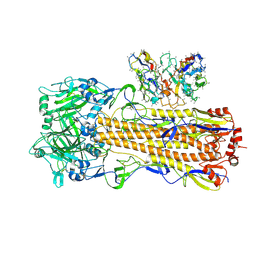 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D28 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT9
 
 | | CryoEM structure of A/Shanghai/1/2013 H7 in complex with polyclonal Fab from mice immunized with H7 stem nanoparticles-28 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT6
 
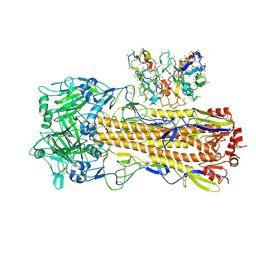 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-15 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D15 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
