6OSO
 
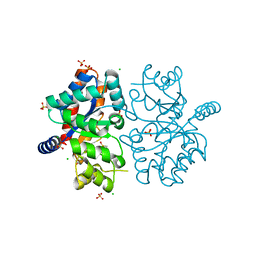 | | The crystal structure of the isolate tryptophan synthase alpha-chain from Salmonella enterica serovar typhimurium at 1.75 Angstrom resolution | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L, Chang, C, Fan, L. | | Deposit date: | 2019-05-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Backbone assignments and conformational dynamics in the S. typhimurium tryptophan synthase alpha-subunit from solution-state NMR.
J.Biomol.Nmr, 74, 2020
|
|
6OUY
 
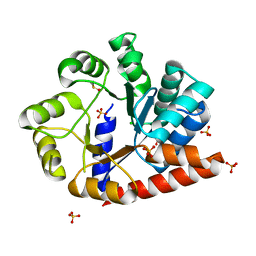 | | The crystal structure of the isolate tryptophan synthase alpha-chain from Salmonella enterica serovar typhimurium at 1.60 Angstrom resolution | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L, Chang, C, Fan, L. | | Deposit date: | 2019-05-06 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Backbone assignments and conformational dynamics in the S. typhimurium tryptophan synthase alpha-subunit from solution-state NMR.
J.Biomol.Nmr, 74, 2020
|
|
6NSL
 
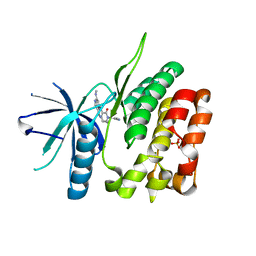 | | CRYSTAL STRUCTURE OF TYROSINE KINASE 2 JH2 (PSEUDO KINASE DOMAIN) COMPLEXED WITH Compound-6c AKA 6-((1-(4-CYANOPHENY L)-2-OXO-1,2-DIHYDRO-3-PYRIDINYL)AMINO)-N-CYCLOPROPYL-8-(M ETHYLAMINO)IMIDAZO[1,2-B]PYRIDAZINE-3-CARBOXAMIDE | | Descriptor: | 6-{[1-(4-cyanophenyl)-2-oxo-1,2-dihydropyridin-3-yl]amino}-N-cyclopropyl-8-(methylamino)imidazo[1,2-b]pyridazine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Muckelbauer, J.M, Khan, J.A. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of Imidazo[1,2-b]pyridazine Derivatives as Potent, Selective, and Orally Active Tyk2 JH2 Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6NZH
 
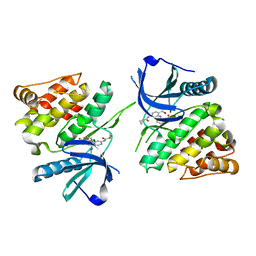 | |
6NZF
 
 | |
6NZE
 
 | |
8K96
 
 | | CryoEM structure of LonC protease hepatmer with Bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K93
 
 | | CryoEM structure of LonC protease S582A open hexamer with lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K97
 
 | | CryoEM structure of LonC protease hexamer with Bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8Y
 
 | | CryoEM structure of LonC heptamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8X
 
 | | CryoEM of LonC open pentamer, apo state | | Descriptor: | endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K92
 
 | | CryoEM structure of LonC S582A hexamer with Lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8V
 
 | | CryoEM structure of LonC protease hepatmer, apo state | | Descriptor: | PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K95
 
 | | CryoEM structure of LonC protease open Hexamer, AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K90
 
 | | CryoEM structure of LonC protease open pentamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8Z
 
 | | CryoEM structure of LonC protease hexamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K91
 
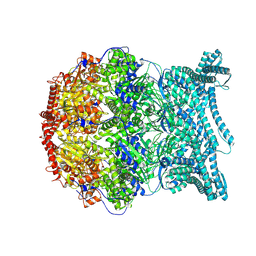 | | CryoEM structure of LonC S582A hepatmer with Lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K94
 
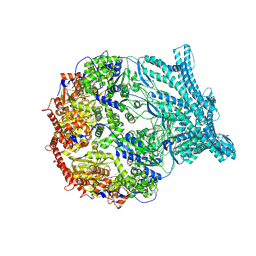 | | CryoEM structure of LonC protease S582A open pentamer with lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8W
 
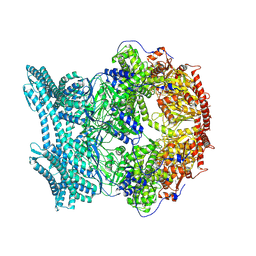 | | CryoEM structure of LonC protease open hexamer, apo state | | Descriptor: | endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
1BVE
 
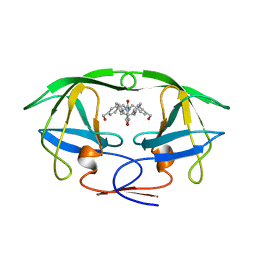 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR, 28 STRUCTURES | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
1BVG
 
 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
7TZP
 
 | | Crystal Structure of Putataive Short-Chain Dehydrogenase/Reductase (FabG) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 in Complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-ACP reductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-16 | | Release date: | 2022-03-02 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7V1Q
 
 | | Leifsonia Alcohol Dehydrogenases LnADH | | Descriptor: | Alcohol Dehydrogenases, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Song, Y, Qu, X. | | Deposit date: | 2021-08-05 | | Release date: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Engineering Leifsonia Alcohol Dehydrogenase for Thermostability and Catalytic Efficiency by Enhancing Subunit Interactions.
Chembiochem, 22, 2021
|
|
7V1R
 
 | | Leifsonia Alcohol Dehydrogenases LnADH | | Descriptor: | Alcohol Dehydrogenases, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Song, Y, Qu, X. | | Deposit date: | 2021-08-05 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Engineering Leifsonia Alcohol Dehydrogenase for Thermostability and Catalytic Efficiency by Enhancing Subunit Interactions.
Chembiochem, 22, 2021
|
|
7K7F
 
 | |
