8AXY
 
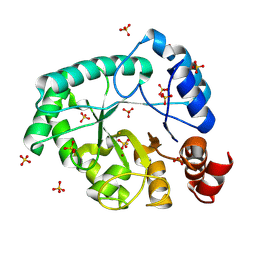 | | Staphylococcus aureus endonuclease IV orthorhombic crystal form | | Descriptor: | FE (III) ION, Probable endonuclease 4, SULFATE ION, ... | | Authors: | Rouvinski, A, Kirillov, S, Saper, M.A, Wiener, R, Isupov, M.N. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Octahedrally coordinated iron in the catalytic site of endonuclease IV from Staphylococcus aureus
To Be Published
|
|
3DV5
 
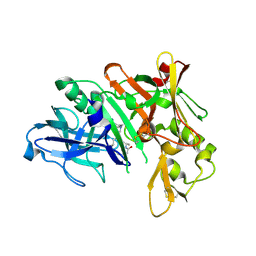 | | Crystal structure of human beta-secretase in complex with NVP-BAV544 | | Descriptor: | (3S,14R,16S)-16-[(1R)-1-hydroxy-2-{[3-(1-methylethyl)benzyl]amino}ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5- dione, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2008-07-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Macrocyclic peptidomimetic beta-secretase (BACE-1) inhibitors with activity in vivo.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5ZLQ
 
 | | Crystal Structure of C1orf123 | | Descriptor: | UPF0587 protein C1orf123, ZINC ION | | Authors: | Furukawa, Y, Lim, C.T, Tosha, T. | | Deposit date: | 2018-03-29 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a novel zinc-binding protein, C1orf123, as an interactor with a heavy metal-associated domain
PLoS ONE, 13, 2018
|
|
3QBH
 
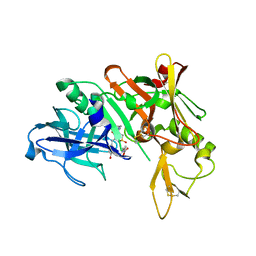 | | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors | | Descriptor: | (4S)-4-(2-hydroxy-5-{[(3S,4S,5R)-4-hydroxy-1,1-dioxido-5-{[3-(propan-2-yl)benzyl]amino}tetrahydro-2H-thiopyran-3-yl]methyl}benzyl)-3-propyl-1,3-oxazolidin-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M. | | Deposit date: | 2011-01-13 | | Release date: | 2011-03-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1B99
 
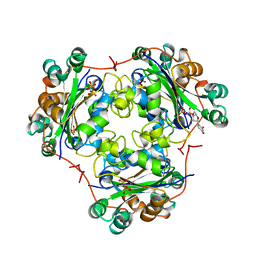 | | 3'-FLUORO-URIDINE DIPHOSPHATE BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 2',3'-DIDEOXY-3'-FLUORO-URIDIDINE-5'-DIPHOSPHATE, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE), PYROPHOSPHATE 2- | | Authors: | Janin, J, Xu, Y. | | Deposit date: | 1999-02-22 | | Release date: | 1999-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of nucleoside diphosphate kinase investigated using nucleotide analogues, viscosity effects, and X-ray crystallography.
Biochemistry, 38, 1999
|
|
2RR4
 
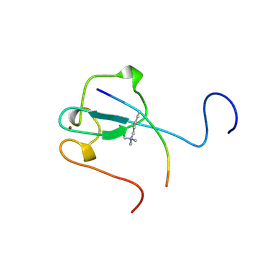 | | Complex structure of the zf-CW domain and the H3K4me3 peptide | | Descriptor: | Histone H3, ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-24 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
2RPJ
 
 | | Solution structure of Fn14 CRD domain | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | He, F, Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-05-19 | | Release date: | 2009-03-24 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain in Fn14, a member of the tumor necrosis factor receptor superfamily
Protein Sci., 18, 2009
|
|
7L4U
 
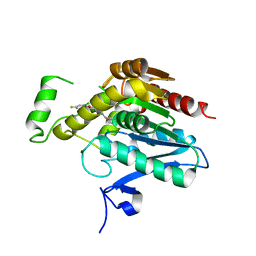 | | Crystal structure of human monoacylglycerol lipase in complex with compound 1h | | Descriptor: | (5S)-5-(3-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidin-1-yl}-3-oxopropyl)pyrrolidin-2-one, CHLORIDE ION, Monoglyceride lipase | | Authors: | Qin, L, Lane, W, Skene, R.J, Dougan, D. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L4W
 
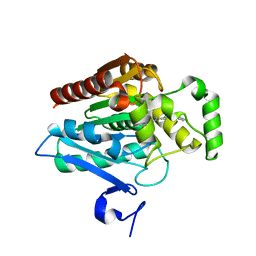 | | Crystal structure of human monoacylglycerol lipase in complex with compound 2d | | Descriptor: | (2s,4R)-2-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidine-1-carbonyl}-7-oxa-5-azaspiro[3.4]octan-6-one, Monoglyceride lipase | | Authors: | Qin, L, Gay, S.C, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L4T
 
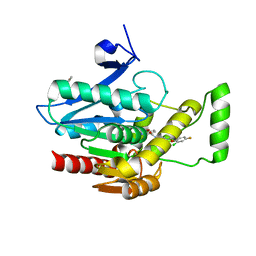 | | Crystal structure of human monoacylglycerol lipase in complex with compound 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidine-1-carbonyl}-2H-1,4-benzoxazin-3(4H)-one, ACETATE ION, ... | | Authors: | Qin, L, Gay, S.C, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7L50
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 4f | | Descriptor: | (2s,4R)-2-{3-[(3-chloro-4-methylphenyl)methoxy]azetidine-1-carbonyl}-7-oxa-5-azaspiro[3.4]octan-6-one, ACETATE ION, Monoglyceride lipase | | Authors: | Qin, L, Lane, W, Skene, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
7FBR
 
 | |
2JNF
 
 | |
7E4D
 
 | | Crystal structure of PlDBR | | Descriptor: | Double Bond Reductase | | Authors: | Sugimoto, K, Senda, M, Senda, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploration and structure-based engineering of alkenal double bond reductases catalyzing the C alpha C beta double bond reduction of coniferaldehyde.
N Biotechnol, 68, 2022
|
|
7FBV
 
 | |
2ZXX
 
 | | Crystal structure of Cdt1/geminin complex | | Descriptor: | DNA replication factor Cdt1, Geminin | | Authors: | Cho, Y, Lee, C, Hong, B.S, Choi, J.M. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibition of the replication licensing factor Cdt1 by geminin
Nature, 430, 2004
|
|
1TIT
 
 | | TITIN, IG REPEAT 27, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TITIN, I27 | | Authors: | Improta, S, Politou, A.S, Pastore, A. | | Deposit date: | 1996-02-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity.
Structure, 4, 1996
|
|
2RSX
 
 | | Solution structure of IseA, an inhibitor protein of DL-endopeptidases from Bacillus subtilis | | Descriptor: | Uncharacterized protein yoeB | | Authors: | Arai, R, Li, H, Tochio, N, Fukui, S, Kobayashi, N, Kitaura, C, Watanabe, S, Kigawa, T, Sekiguchi, J. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of IseA, an Inhibitor Protein of DL-Endopeptidases from Bacillus subtilis, Reveals a Novel Fold with a Characteristic Inhibitory Loop
J.Biol.Chem., 287, 2012
|
|
7XGR
 
 | |
7XDT
 
 | |
2V34
 
 | | IspE in complex with cytidine and ligand | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, CHLORIDE ION, ... | | Authors: | Sgraja, T, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Aquifex Aeolicus 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase -Ligand Recognition in a Template for Antimicrobial Drug Discovery.
FEBS J., 275, 2008
|
|
8AN8
 
 | | Crystal structure of wild-type c-MET bound by compound 7. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
8ANS
 
 | | Crystal structure of D1228V c-MET bound by compound 1. | | Descriptor: | 3-[bis(fluoranyl)methyl]-~{N}-methyl-~{N}-[(1~{R})-8-methyl-5-(3-methyl-1~{H}-indazol-6-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]pyridine-2-carboxamide, GLYCEROL, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2022-08-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of a selective c-MET inhibitor with a novel binding mode.
Bioorg.Med.Chem.Lett., 75, 2022
|
|
7UOC
 
 | | Crystal structure of Orobanche minor KAI2d4 | | Descriptor: | CHLORIDE ION, KAI2d4 | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Divergent Clade KAI2 Protein in the Root Parasitic Plant Orobanche minor Is a Highly Sensitive Strigolactone Receptor and Is Involved in the Perception of Sesquiterpene Lactones.
Plant Cell.Physiol., 64, 2023
|
|
2V2Z
 
 | | IspE in complex with ADP and CDPME | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sgraja, T, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-06-08 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of Aquifex Aeolicus 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase -Ligand Recognition in a Template for Antimicrobial Drug Discovery.
FEBS J., 275, 2008
|
|
