8QFH
 
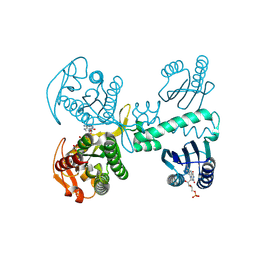 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFJ
 
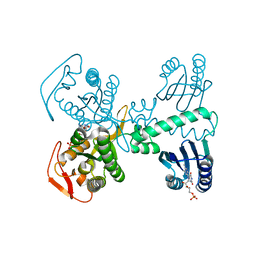 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC after blue light excitation at 2.3 us delay | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFI
 
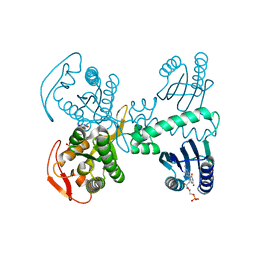 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC after blue light excitation at 1.8 us delay | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Sato, T, Round, A, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
1LFW
 
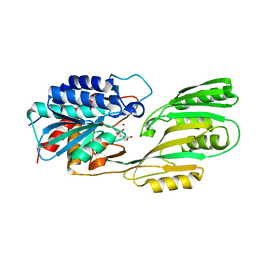 | | Crystal structure of pepV | | Descriptor: | 3-[(1-AMINO-2-CARBOXY-ETHYL)-HYDROXY-PHOSPHINOYL]-2-METHYL-PROPIONIC ACID, ZINC ION, pepV | | Authors: | Jozic, D, Bourenkow, G, Bartunik, H, Scholze, H, Dive, V, Henrich, B, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Dinuclear Zinc Aminopeptidase PepV from Lactobacillus delbrueckii Unravels Its Preference for Dipeptides
Structure, 10, 2002
|
|
1JFD
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Oganesyan, V, Avaeva, S.M, Huber, R, Harutyunyan, E.H. | | Deposit date: | 1997-05-31 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Escherichia coli inorganic pyrophosphatase complexed with SO4(2-). Ligand-induced molecular asymmetry.
FEBS Lett., 410, 1997
|
|
1LTO
 
 | | Human alpha1-tryptase | | Descriptor: | alpha tryptase I | | Authors: | Marquardt, U, Zettl, F, Huber, R, Bode, W, Sommerhoff, C.P. | | Deposit date: | 2002-05-20 | | Release date: | 2003-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of Human alpha1-Tryptase Reveals a Blocked Substrate-binding Region
J.MOL.BIOL., 321, 2002
|
|
2FWH
 
 | | atomic resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form at pH7) | | Descriptor: | DI(HYDROXYETHYL)ETHER, IODIDE ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWF
 
 | | high resolution crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (reduced form) | | Descriptor: | IODIDE ION, SODIUM ION, Thiol:disulfide interchange protein dsbD | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2FWE
 
 | | crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (oxidized form) | | Descriptor: | IODIDE ION, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
1SNN
 
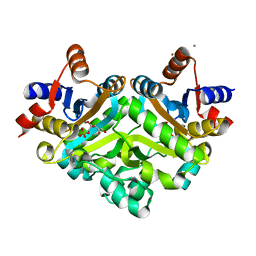 | | 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CALCIUM ION, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Steinbacher, S, Schiffmann, S, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2004-03-11 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metal sites in 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with the substrate ribulose 5-phosphate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8OQJ
 
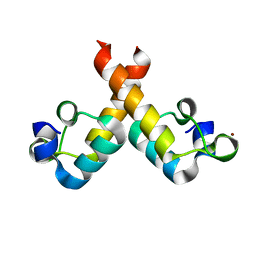 | |
8ORB
 
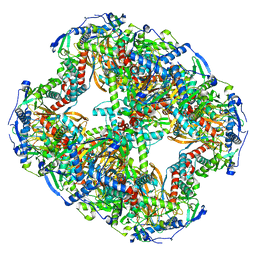 | |
8OSY
 
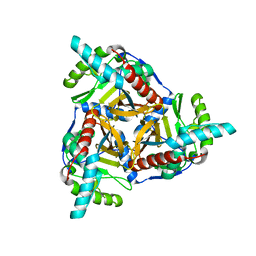 | |
6Y7S
 
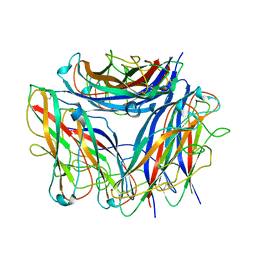 | | 2.85 A cryo-EM structure of the in vivo assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Hospenthal, M, Waksman, G, Glockshuber, R. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
8PSV
 
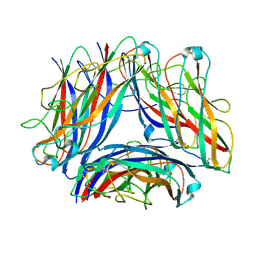 | | 2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M, Zyla, D, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
8PTU
 
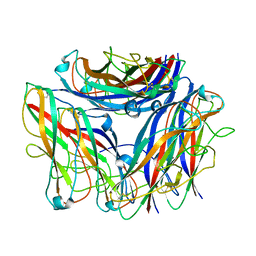 | | 2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Hospenthal, M, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
7OT4
 
 | |
8OET
 
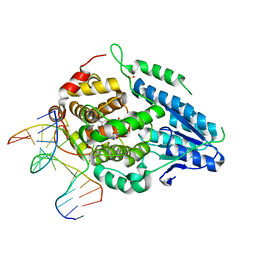 | | SFX structure of the class II photolyase complexed with a thymine dimer | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, DNA (14-mer), Deoxyribodipyrimidine photo-lyase, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-03-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
5IQN
 
 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitution Q134E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_SRIRIRGYVR | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Protein FimF, ... | | Authors: | Giese, C, Eras, J, Kern, A, Scharer, M.A, Capitani, G, Glockshuber, R. | | Deposit date: | 2016-03-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8OYC
 
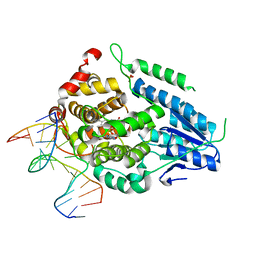 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (100 microsecond timpeoint) | | Descriptor: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
4X43
 
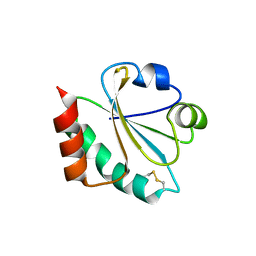 | |
4XO9
 
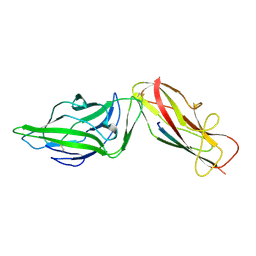 | | Crystal structure of a FimH*DsG complex from E.coli K12 in space group C2 | | Descriptor: | Minor component of type 1 fimbriae, Protein FimH | | Authors: | Jakob, R.P, Eras, J, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
5IQM
 
 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitution Q134E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_T4R-T6R-D13N | | Descriptor: | COBALT (II) ION, Protein FimF, Protein FimG | | Authors: | Giese, C, Eras, J, Kern, A, Scharer, M.A, Capitani, G, Glockshuber, R. | | Deposit date: | 2016-03-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4XO8
 
 | | Crystal structure of the FimH lectin domain from E.coli K12 in complex with heptyl alpha-D-mannopyrannoside | | Descriptor: | Protein FimH, heptyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Eras, J, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
