5XUS
 
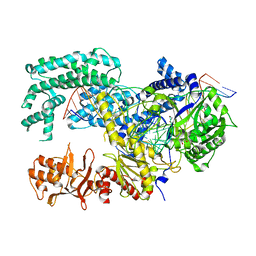 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TTTA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
5XH6
 
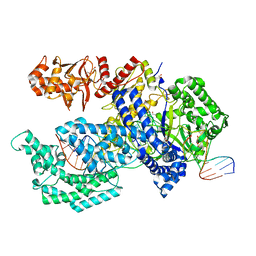 | | Crystal structure of the Acidaminococcus sp. BV3L6 Cpf1 RVR variant in complex with crRNA and target DNA (TATA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CRISPR-associated endonuclease Cpf1, ... | | Authors: | Nishimasu, H, Yamano, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Altered PAM Recognition by Engineered CRISPR-Cpf1
Mol. Cell, 67, 2017
|
|
5XUU
 
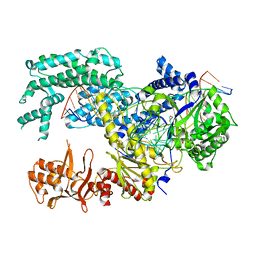 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
6AEK
 
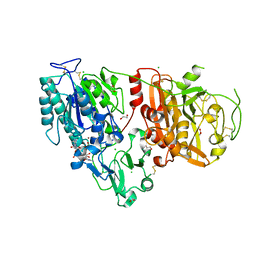 | | Crystal structure of ENPP1 in complex with pApG | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
6AEL
 
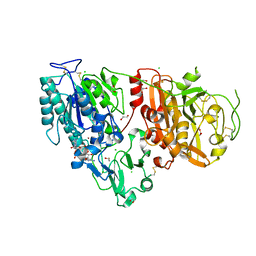 | | Crystal structure of ENPP1 in complex with 3'3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, ... | | Authors: | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
2JYG
 
 | |
2JYL
 
 | |
2DR9
 
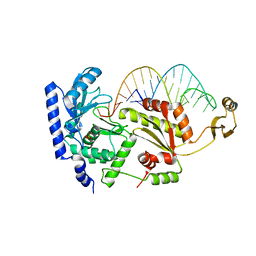 | | Complex structure of CCA-adding enzyme with tRNAminiDCC | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (34-MER) | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DRA
 
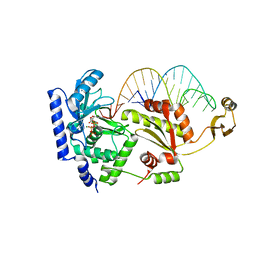 | | Complex structure of CCA-adding enzyme with tRNAminiDCC and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CCA-adding enzyme, MAGNESIUM ION, ... | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DR5
 
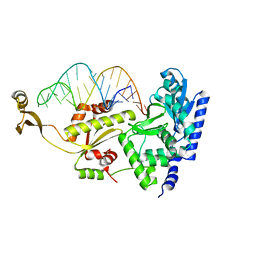 | | Complex structure of CCA adding enzyme with mini-helix lacking CCA | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (32-MER) | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DRB
 
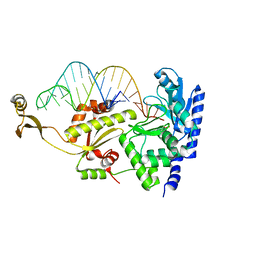 | | Complex structure of CCA-adding enzyme with tRNAminiCCA | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (35-MER) | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DR7
 
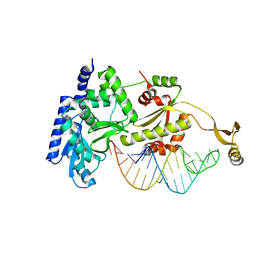 | | Complex structure of CCA-adding enzyme with tRNAminiDC | | Descriptor: | CCA-adding enzyme, SULFATE ION, tRNA (33-MER) | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-06-08 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2DVI
 
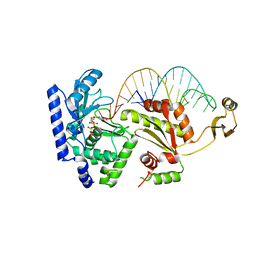 | | Complex structure of CCA-adding enzyme, mini-DCC and CTP | | Descriptor: | CCA-adding enzyme, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tomita, K, Ishitani, R, Fukai, S, Nureki, O. | | Deposit date: | 2006-07-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Complete crystallographic analysis of the dynamics of CCA sequence addition
Nature, 443, 2006
|
|
2EL9
 
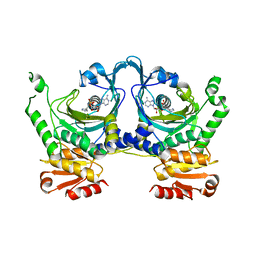 | | Crystal structure of E.coli Histidyl-tRNA synthetase complexed with a histidyl-adenylate analogue | | Descriptor: | 5'-O-[(L-HISTIDYLAMINO)SULFONYL]ADENOSINE, Histidyl-tRNA synthetase | | Authors: | Yanagisawa, T, Si, S.Y, Matsuno, M, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of E.coli Histidyl-tRNA synthetase complexed with a histidyl-adenylate analogue
To be published
|
|
2EL7
 
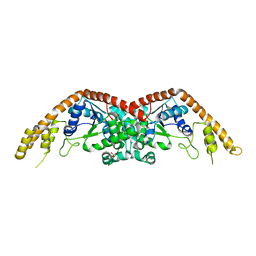 | | Crystal structure of Tryptophanyl-tRNA synthetase from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, Tryptophanyl-tRNA synthetase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Thermus thermophilus
To be published
|
|
6AID
 
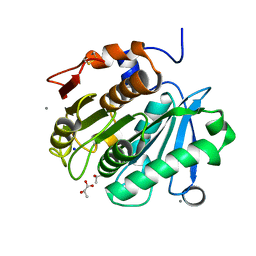 | | Structural insights into the unique polylactate degrading mechanism of Thermobifida alba cutinase | | Descriptor: | CALCIUM ION, Esterase, LACTIC ACID, ... | | Authors: | Kitadokoro, K, Kakara, M, Matsui, S, Osokoshi, R, Thumarat, U, Kawai, F, Kamitani, S. | | Deposit date: | 2018-08-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase.
Febs J., 286, 2019
|
|
2E3C
 
 | |
3WNC
 
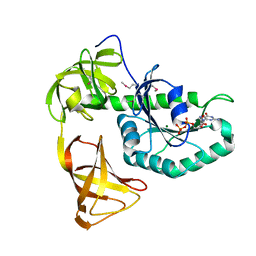 | | Crystal structure of EF-Pyl in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
3WNB
 
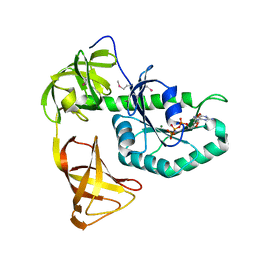 | | Crystal structure of EF-Pyl in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
3WND
 
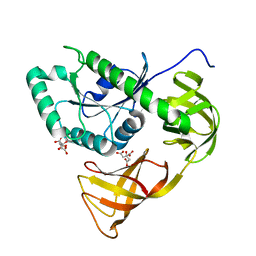 | | Crystal structure of EF-Pyl | | Descriptor: | CITRIC ACID, Protein translation elongation factor 1A | | Authors: | Yanagisawa, T, Ishii, R, Fukunaga, R, Sengoku, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2013-12-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A SelB/EF-Tu/aIF2 gamma-like protein from Methanosarcina mazei in the GTP-bound form binds cysteinyl-tRNA(Cys.).
J. Struct. Funct. Genomics, 16, 2015
|
|
3UG9
 
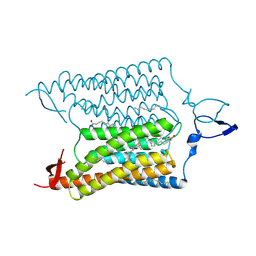 | | Crystal Structure of the Closed State of Channelrhodopsin | | Descriptor: | Archaeal-type opsin 1, Archaeal-type opsin 2, OLEIC ACID, ... | | Authors: | Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2011-11-02 | | Release date: | 2012-01-25 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the channelrhodopsin light-gated cation channel
Nature, 482, 2012
|
|
3VVZ
 
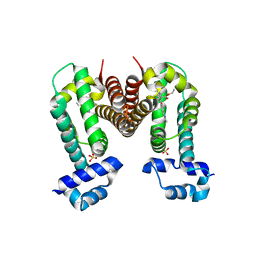 | | Crystal Strucuture of The Rhodamine 6G-Bound Form of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | Descriptor: | Putative regulatory protein, RHODAMINE 6G, SULFATE ION | | Authors: | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
3VVX
 
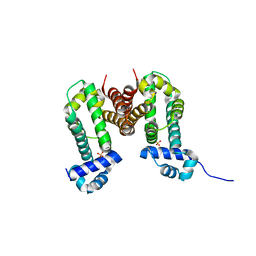 | | Crystal Strucuture of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | Descriptor: | Putative regulatory protein, SULFATE ION | | Authors: | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
3VVY
 
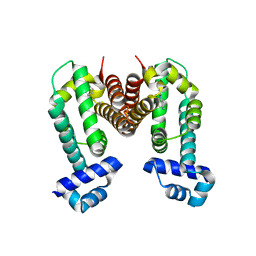 | | Crystal Strucuture of The Ethidium-Bound Form of RamR (Transcriptional Regurator of TetR Family) from Salmonella Typhimurium | | Descriptor: | ETHIDIUM, Putative regulatory protein | | Authors: | Sakurai, K, Nikaido, E, Nakashima, R, Yamasaki, S, Yamaguchi, A, Nishino, K. | | Deposit date: | 2012-07-30 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The crystal structure of multidrug-resistance regulator RamR with multiple drugs
Nat Commun, 4, 2013
|
|
3VUW
 
 | | Crystal structure of A20 ZF7 in complex with linear ubiquitin, form I | | Descriptor: | POTASSIUM ION, Polyubiquitin-C, Tumor necrosis factor alpha-induced protein 3, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation
Embo J., 31, 2012
|
|
