2KWK
 
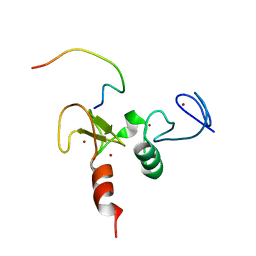 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3b bound to a H3 peptide wild type | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
2L5D
 
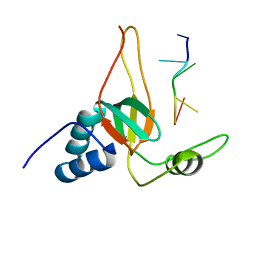 | | Solution Structures of human PIWI-like 1 PAZ domain with ssRNA (5'-pUGACA) | | Descriptor: | 5'-R(*UP*GP*AP*CP*A)-3', Piwi-like protein 1 | | Authors: | Zeng, L, Zhang, Q, Yan, K, Zhou, M. | | Deposit date: | 2010-10-29 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into piRNA recognition by the human PIWI-like 1 PAZ domain.
Proteins, 79, 2011
|
|
1QHT
 
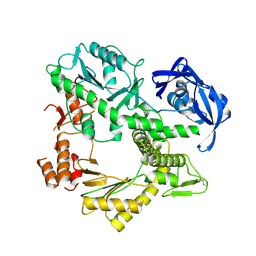 | |
2NCZ
 
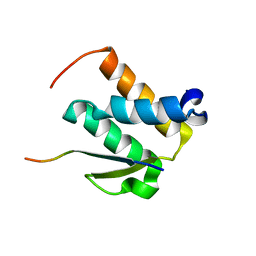 | |
2ND1
 
 | |
2NDF
 
 | |
2ND0
 
 | |
2NDG
 
 | |
2L5C
 
 | |
2KWJ
 
 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3 bound to a histone peptide containing acetylation at lysine 14 | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
2KFT
 
 | |
1YQH
 
 | |
1YB4
 
 | | Crystal Structure of the Tartronic Semialdehyde Reductase from Salmonella typhimurium LT2 | | Descriptor: | tartronic semialdehyde reductase | | Authors: | Kim, Y, Wu, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of GarR-tartronate semialdehyde reductase from Salmonella typhimurium.
J Struct Funct Genomics, 10, 2009
|
|
3DB3
 
 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 in complex with trimethylated histone H3-K9 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Trimethylated histone H3-K9 peptide | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
3DB4
 
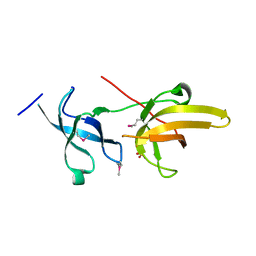 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
3MW6
 
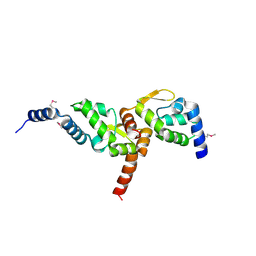 | | Crystal structure of NMB1681 from Neisseria meningitidis MC58, a FinO-like RNA chaperone | | Descriptor: | GLYCEROL, uncharacterized protein NMB1681 | | Authors: | Tan, K, Zhou, M, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | N. meningitidis 1681 is a member of the FinO family of RNA chaperones.
Rna Biol., 7, 2010
|
|
3ZJA
 
 | |
3ZK0
 
 | |
3N5C
 
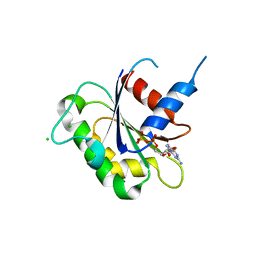 | | Crystal Structure of Arf6DELTA13 complexed with GDP | | Descriptor: | ADP-ribosylation factor 6, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Aizel, K, Biou, V, Cherfils, J. | | Deposit date: | 2010-05-25 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SAXS and X-ray crystallography suggest an unfolding model for the GDP/GTP conformational switch of the small GTPase Arf6.
J.Mol.Biol., 402, 2010
|
|
2G17
 
 | |
2L3R
 
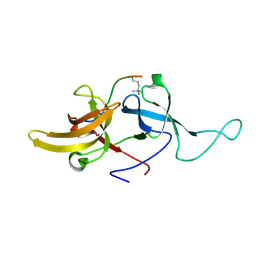 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3 | | Authors: | Nady, N, Lemak, A, Fares, C, Gutmanas, A, Avvakumov, G, Xue, S, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-21 | | Release date: | 2011-04-13 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Recognition of Multivalent Histone States Associated with Heterochromatin by UHRF1 Protein.
J.Biol.Chem., 286, 2011
|
|
1AWV
 
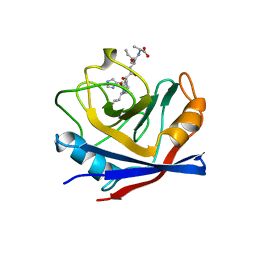 | | CYPA COMPLEXED WITH HVGPIA | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-05 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1AWU
 
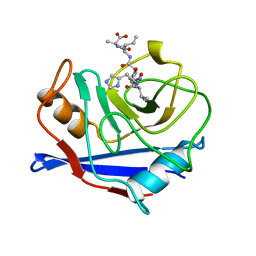 | | CYPA COMPLEXED WITH HVGPIA (PSEUDO-SYMMETRIC MONOMER) | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-05 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1AWT
 
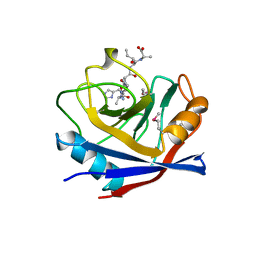 | | SECYPA COMPLEXED WITH HAGPIA | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-05 | | Release date: | 1998-03-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1AWS
 
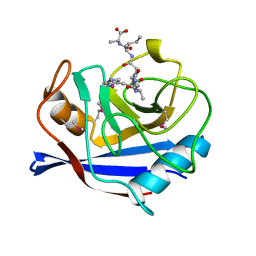 | | SECYPA COMPLEXED WITH HAGPIA (PSEUDO-SYMMETRIC MONOMER) | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-04 | | Release date: | 1998-03-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
