5FH8
 
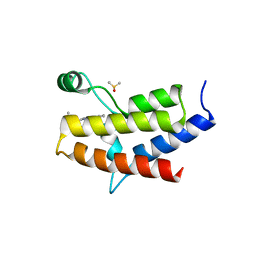 | | Crystal structure of the fifth bromodomain of human PB1 in complex with compound 28 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-3-(2-ethylbutyl)-4~{H}-pyrrolo[1,2-a]quinazolin-5-one, DIMETHYL SULFOXIDE, ... | | Authors: | Tallant, C, Sutherell, C.L, Siejka, P, Sorrell, F.J, Krojer, T, Picaud, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Ley, S.V, Knapp, S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Development of 2,3-Dihydropyrrolo[1,2-a]quinazolin-5(1H)-one Inhibitors Targeting Bromodomains within the Switch/Sucrose Nonfermenting Complex.
J.Med.Chem., 59, 2016
|
|
6N3F
 
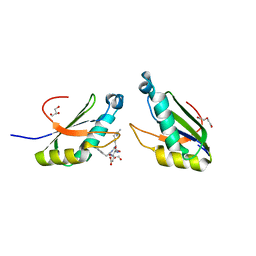 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to SF3b1 ULM5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HIV Tat-specific factor 1, ... | | Authors: | Leach, J.R, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
6N2O
 
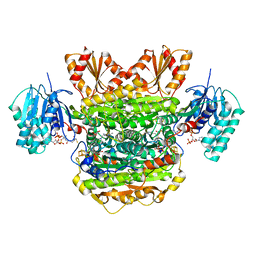 | |
6N3E
 
 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif bound to U2AF ligand motif 4 | | Descriptor: | FORMIC ACID, GLYCEROL, HIV Tat-specific factor 1, ... | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
6MWX
 
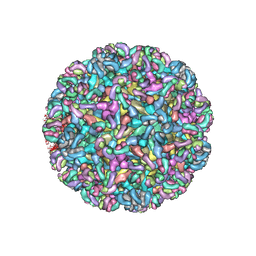 | | CryoEM structure of Chimeric Eastern Equine Encephalitis Virus with Fab of EEEV-69 Antibody | | Descriptor: | E1, E2, EEEV-69 antibody heavy chain, ... | | Authors: | Hasan, S.S, Sun, C, Kim, A.S, Watanabe, Y, Chen, C.L, Klose, T, Buda, G, Crispin, M, Diamond, M.S, Klimstra, W.B, Rossmann, M.G. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM Structures of Eastern Equine Encephalitis Virus Reveal Mechanisms of Virus Disassembly and Antibody Neutralization.
Cell Rep, 25, 2018
|
|
5G2X
 
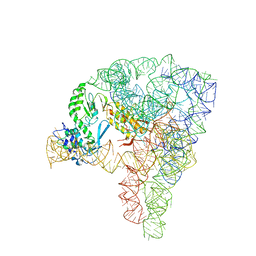 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | 5'-R(*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*CP)-3', GROUP II INTRON, GROUP II INTRON-ENCODED PROTEIN LTRA | | Authors: | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5FBK
 
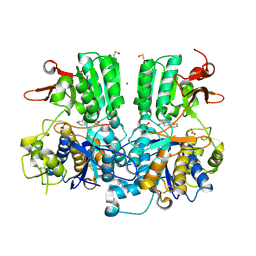 | | Crystal structure of the extracellular domain of human calcium sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
6N3D
 
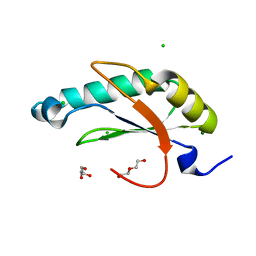 | | Structure of HIV Tat-specific factor 1 U2AF Homology Motif (APO-State) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The pre-mRNA splicing and transcription factor Tat-SF1 is a functional partner of the spliceosome SF3b1 subunit via a U2AF homology motif interface.
J. Biol. Chem., 294, 2019
|
|
1FNN
 
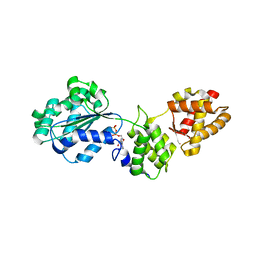 | | CRYSTAL STRUCTURE OF CDC6P FROM PYROBACULUM AEROPHILUM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6, MAGNESIUM ION | | Authors: | Liu, J, Smith, C.L, DeRyckere, D, DeAngelis, K, Martin, G.S, Berger, J.M. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control.
Mol.Cell, 6, 2000
|
|
7WQA
 
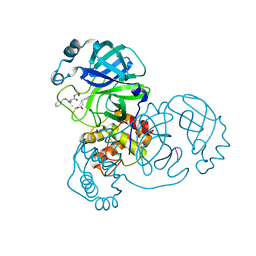 | | SARS-CoV-2 main protease in complex with Z-VAD-FMK | | Descriptor: | 3C-like proteinase, Z-VAD(OMe)-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 main protease in complex with Z-VAD-FMK
To Be Published
|
|
7WQ9
 
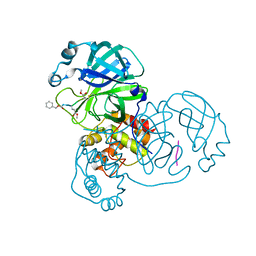 | | Crystal structure of SARS-CoV-2 main protease in complex with Z-IETD-FMK | | Descriptor: | 3C-like proteinase, Z-IETD-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with Z-IETD-FMK
To Be Published
|
|
7WQ8
 
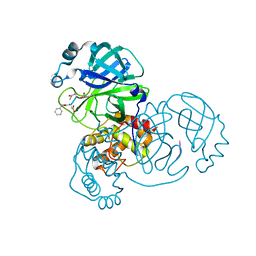 | | Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMK | | Descriptor: | 3C-like proteinase, Z-DEVD-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMK
To Be Published
|
|
7WQB
 
 | | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132
To Be Published
|
|
7WQK
 
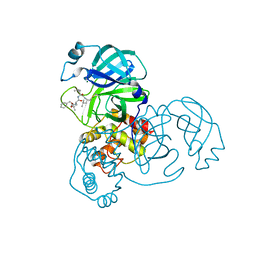 | | wild-type SARS-CoV-2 main protease in complex with MG-132 | | Descriptor: | 3C-like proteinase, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | wild-type SARS-CoV-2 main protease in complex with MG-132
To Be Published
|
|
6ND3
 
 | |
5G2Y
 
 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | GROUP II INTRON | | Authors: | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5FH7
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with compound 18 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-3-[(dimethylamino)methyl]-4~{H}-pyrrolo[1,2-a]quinazolin-5-one, Protein polybromo-1 | | Authors: | Tallant, C, Sutherell, C.L, Siejka, P, Sorrell, F.J, Krojer, T, Picaud, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Ley, S.V, Knapp, S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Identification and Development of 2,3-Dihydropyrrolo[1,2-a]quinazolin-5(1H)-one Inhibitors Targeting Bromodomains within the Switch/Sucrose Nonfermenting Complex.
J.Med.Chem., 59, 2016
|
|
5FAV
 
 | | E491Q mutant of choline TMA-lyase | | Descriptor: | Choline trimethylamine-lyase, MALONATE ION, SODIUM ION | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2015-12-12 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Basis of C-N Bond Cleavage by the Glycyl Radical Enzyme Choline Trimethylamine-Lyase.
Cell Chem Biol, 23, 2016
|
|
1ID3
 
 | | CRYSTAL STRUCTURE OF THE YEAST NUCLEOSOME CORE PARTICLE REVEALS FUNDAMENTAL DIFFERENCES IN INTER-NUCLEOSOME INTERACTIONS | | Descriptor: | HISTONE H2A.1, HISTONE H2B.2, HISTONE H3, ... | | Authors: | White, C.L, Suto, R.K, Luger, K. | | Deposit date: | 2001-04-03 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions.
EMBO J., 20, 2001
|
|
1G89
 
 | |
1I7E
 
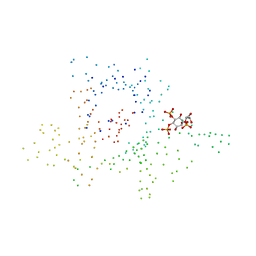 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | Descriptor: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | Authors: | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
1IW2
 
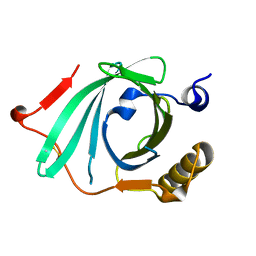 | | X-ray structure of Human Complement Protein C8gamma at pH=7.O | | Descriptor: | Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
5T3W
 
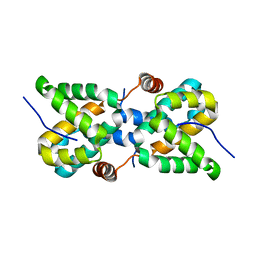 | |
5T3T
 
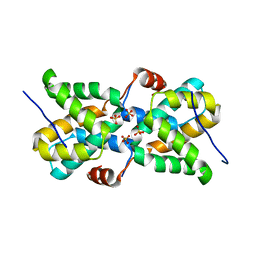 | | Ebola virus VP30 CTD bound to nucleoprotein | | Descriptor: | Fusion protein of Nucleoprotein and Minor nucleoprotein VP30, SULFATE ION | | Authors: | Kirchdoerfer, R.N, Moyer, C.L, Abelson, D.M, Saphire, E.O. | | Deposit date: | 2016-08-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Ebola Virus VP30-NP Interaction Is a Regulator of Viral RNA Synthesis.
Plos Pathog., 12, 2016
|
|
5T7Z
 
 | | Monoclinic crystal form of the EpoB NRPS cyclization-docking bidomain from Sorangium cellulosum | | Descriptor: | EpoB | | Authors: | Dowling, D.P, Kung, Y, Croft, A.K, Taghizadeh, K, Kelly, W.L, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2016-09-06 | | Release date: | 2016-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural elements of an NRPS cyclization domain and its intermodule docking domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
