2HRN
 
 | | Solution Structure of Cu(I) P174L-HSco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Leontari, I, Martinelli, M, Palumaa, P, Sillard, R, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human Sco1 functional studies and pathological implications of the P174L mutant.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8EX3
 
 | | Plasmodium falciparum M1 in complex with inhibitor 9aa | | Descriptor: | GLYCEROL, M1 family aminopeptidase, MAGNESIUM ION, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
8EYD
 
 | | Plasmodium falciparum M1 in complex with inhibitor 15ah | | Descriptor: | GLYCEROL, M1 family aminopeptidase, N-[(1R)-1-(4-bromophenyl)-2-(hydroxyamino)-2-oxoethyl]-N~2~-(4-fluorophenyl)glycinamide, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
4U3H
 
 | | Crystal structure of FN3con | | Descriptor: | FN3con | | Authors: | Porebski, B.T, McGowan, S, Buckle, A.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and dynamic properties that govern the stability of an engineered fibronectin type III domain.
Protein Eng.Des.Sel., 28, 2015
|
|
6DRZ
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N-[(2S)-1-hydroxybutan-2-yl]-1,6-dimethyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRY
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRX
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, BRIL chimera, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DS0
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (1S)-1-[(2-chloro-3,4-dimethoxyphenyl)methyl]-6-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1JQ0
 
 | | Mutation that destabilize the gp41 core: determinants for stabilizing the SIV/CPmac envelope glycoprotein complex. Mutant structure. | | Descriptor: | gp41 envelope protein | | Authors: | Liu, J, Wang, S, LaBranche, C.C, Hoxie, J.A, Lu, M. | | Deposit date: | 2001-08-03 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutations that destabilize the gp41 core are determinants for stabilizing the simian immunodeficiency virus-CPmac envelope glycoprotein complex.
J.Biol.Chem., 277, 2002
|
|
5XBL
 
 | | Structure of nuclease in complex with associated protein | | Descriptor: | Associated protein, CRISPR-associated endonuclease Cas9/Csn1, RNA (98-MER) | | Authors: | Dong, D, Guo, M, Wang, S, Zhu, Y, Huang, Z. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein
Nature, 546, 2017
|
|
4Y21
 
 | | Crystal Structure of Munc13-1 MUN domain | | Descriptor: | Protein unc-13 homolog A | | Authors: | Yang, X.Y, Wang, S, Sheng, Y, Zhang, M, Zou, W.J, Wu, L.J, Kang, L.J, Rizo, J, Zhang, R.G, Xu, T, Ma, C. | | Deposit date: | 2015-02-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XNH
 
 | |
4XPD
 
 | |
4Y49
 
 | |
4YRY
 
 | | Insights into flavin-based electron bifurcation via the NADH-dependent reduced ferredoxin-NADP oxidoreductase structure | | Descriptor: | Dihydroorotate dehydrogenase B (NAD(+)), electron transfer subunit homolog, Dihydropyrimidine dehydrogenase subunit A, ... | | Authors: | Ermler, U, Thauer, R.K, Demmer, J.K, Huang, H, Wang, S, Demmer, U. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into Flavin-based Electron Bifurcation via the NADH-dependent Reduced Ferredoxin:NADP Oxidoreductase Structure.
J.Biol.Chem., 290, 2015
|
|
4X2T
 
 | | X-ray crystal structure of the orally available aminopeptidase inhibitor, Tosedostat, bound to the M17 Leucyl Aminopeptidase from P. falciparum | | Descriptor: | (2S)-({(2R)-2-[(1S)-1-hydroxy-2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}amino)(phenyl)ethanoic acid, CARBONATE ION, M17 leucyl aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | X-ray crystal structures of an orally available aminopeptidase inhibitor, Tosedostat, bound to anti-malarial drug targets PfA-M1 and PfA-M17.
Proteins, 83, 2015
|
|
1AOC
 
 | | JAPANESE HORSESHOE CRAB COAGULOGEN | | Descriptor: | COAGULOGEN, SULFATE ION | | Authors: | Bergner, A, Oganessyan, V, Muta, T, Iwanaga, S, Typke, D, Huber, R, Bode, W. | | Deposit date: | 1996-11-28 | | Release date: | 1997-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a coagulogen, the clotting protein from horseshoe crab: a structural homologue of nerve growth factor.
EMBO J., 15, 1996
|
|
5WSO
 
 | |
4YLF
 
 | | Insights into flavin-based electron bifurcation via the NADH-dependent reduced ferredoxin-NADP oxidoreductase structure | | Descriptor: | Dihydroorotate dehydrogenase B (NAD(+)), electron transfer subunit homolog, Dihydropyrimidine dehydrogenase subunit A, ... | | Authors: | Ermler, U, Thauer, R.K, Demmer, J.K, Huang, H, Wang, S, Demmer, U. | | Deposit date: | 2015-03-05 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insights into Flavin-based Electron Bifurcation via the NADH-dependent Reduced Ferredoxin:NADP Oxidoreductase Structure.
J.Biol.Chem., 290, 2015
|
|
4YTV
 
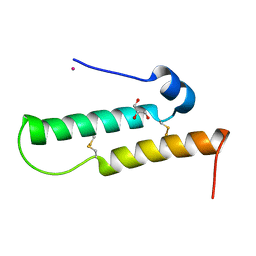 | | Crystal structure of Mdm35 | | Descriptor: | COBALT (II) ION, GLYCEROL, Mitochondrial distribution and morphology protein 35 | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
4YTW
 
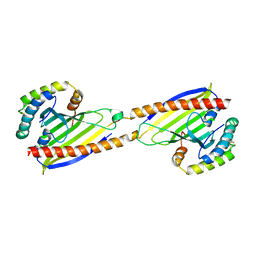 | | Crystal structure of Ups1-Mdm35 complex | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Watanabe, Y, Tamura, Y, Kawano, S, Endo, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic insights into phospholipid transfer by Ups1-Mdm35 in mitochondria.
Nat Commun, 6, 2015
|
|
4X2U
 
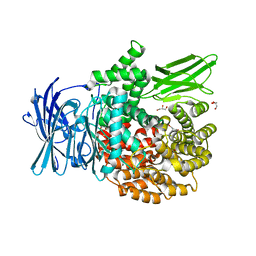 | | X-ray crystal structure of the orally available aminopeptidase inhibitor, Tosedostat, bound to the M1 Alanyl Aminopeptidase from P. falciparum | | Descriptor: | (2S)-({(2R)-2-[(1S)-1-hydroxy-2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}amino)(phenyl)ethanoic acid, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structures of an orally available aminopeptidase inhibitor, Tosedostat, bound to anti-malarial drug targets PfA-M1 and PfA-M17.
Proteins, 83, 2015
|
|
8T6H
 
 | |
8T83
 
 | |
8T7P
 
 | | X-ray crystal structure of PfA-M1(M462S) | | Descriptor: | Aminopeptidase N, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yang, W, Drinkwater, N, Webb, C.T, McGowan, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational dynamics of the Plasmodium falciparum M1 aminopeptidase.
To Be Published
|
|
