5VBU
 
 | |
6DJ4
 
 | |
6KI2
 
 | |
6KI1
 
 | |
6LPV
 
 | |
6LPW
 
 | |
5XN6
 
 | | Heterodimer crystal structure of geranylgeranyl diphosphate synthases 1 with GGPPS Recruiting Protein(OsGRP) from Oryza sativa | | Descriptor: | Os02g0668100 protein, Os07g0580900 protein | | Authors: | Wang, C, Zhou, F, Lu, S, Zhang, P. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.598 Å) | | Cite: | A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7YCV
 
 | |
7YCU
 
 | |
7YCW
 
 | |
7YCS
 
 | |
7EQT
 
 | |
7EQS
 
 | |
7EQW
 
 | |
7ER1
 
 | |
7ER0
 
 | |
8K48
 
 | | LTD of arabidopsis thaliana | | Descriptor: | Protein LHCP TRANSLOCATION DEFECT | | Authors: | Wang, C, Xu, X.M. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Arabidopsis thaliana LTD
To Be Published
|
|
7Y4T
 
 | | Crystal structure of cMET kinase domain bound by compound 9I | | Descriptor: | 2-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-(3-nitrophenyl)pyridazin-3-one, Hepatocyte growth factor receptor | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
7Y4U
 
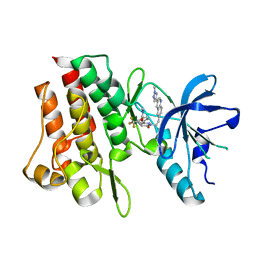 | | Crystal structure of cMET kinase domain bound by compound 9Y | | Descriptor: | Hepatocyte growth factor receptor, ~{N}-methyl-4-[1-[2-[3-(1-methylpyrazol-4-yl)quinolin-6-yl]ethyl]-6-oxidanylidene-pyridazin-3-yl]-2-(trifluoromethyl)benzamide | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
5PZM
 
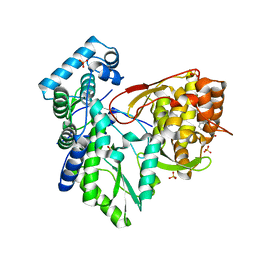 | |
5PZN
 
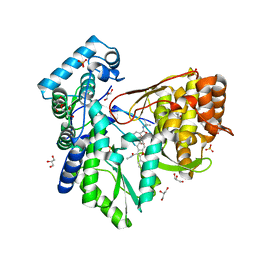 | |
6N6B
 
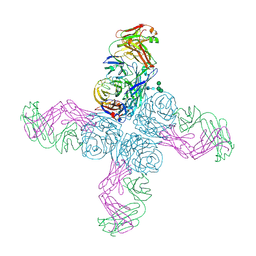 | |
7V6F
 
 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase complexed with G3P | | Descriptor: | Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ZINC ION | | Authors: | Hongxuan, C, Huang, Y, Han, C, Chen, W, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
8K2S
 
 | |
8K2R
 
 | |
