2VTP
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, {[(2,6-difluorophenyl)carbonyl]amino}-N-(4-fluorophenyl)-1H-pyrazole-3-carboxamide | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2J14
 
 | | 3,4,5-Trisubstituted Isoxazoles as Novel PPARdelta Agonists: Part2 | | Descriptor: | (3-{4-[2-(2,4-DICHLORO-PHENOXY)-ETHYLCARBAMOYL]-5-PHENYL-ISOXAZOL-3-YL}-PHENYL)-ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR DELTA | | Authors: | Epple, R, Azimioara, M, Russo, R, Xie, Y, Wang, X, Cow, C, Wityak, J, Karanewsky, D, Bursulaya, B, Kreusch, A, Tuntland, T, Gerken, A, Iskandar, M, Saez, E, Seidel, H.M, Tian, S.S. | | Deposit date: | 2006-08-08 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3,4,5-Trisubstituted Isoxazoles as Novel Ppardelta Agonists. Part 2
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2IWE
 
 | | Structure of a cavity mutant (H117G) of Pseudomonas aeruginosa azurin | | Descriptor: | 1,1'-HEXANE-1,6-DIYLBIS(1H-IMIDAZOLE), AZURIN, ZINC ION | | Authors: | De Jongh, T.E, Van Roon, A.M.M, Prudencio, M, Ubbink, M, Canters, G.W. | | Deposit date: | 2006-06-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Click-Chemistry with an Active Site Variant of Azurin
Eur.J.Inorg.Chem., 2006, 2006
|
|
2IYN
 
 | | The co-factor-induced pre-active conformation in PhoB | | Descriptor: | MAGNESIUM ION, PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Sola, M, Drew, D.L, Blanco, A.G, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 2006-07-19 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Cofactor-Induced Pre-Active Conformation in Phob.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IGQ
 
 | | Human euchromatic histone methyltransferase 1 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, euchromatic histone methyltransferase 1 | | Authors: | Min, J, Wu, H, Antoshenko, T, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-23 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
3ZNS
 
 | | HDAC7 bound with TFMO inhibitor tmp942 | | Descriptor: | HISTONE DEACETYLASE 7, N-{[1-methyl-4-(4-phenyl-1,3-thiazol-2-yl)piperidin-4-yl]methyl}-3-[5-(trifluoromethyl)-1,2,4-oxadiazol-3-yl]benzamide, POTASSIUM ION, ... | | Authors: | Lobera, M, Madauss, K.P, Pohlhaus, D.T, Trump, R.P, Nolan, M.A. | | Deposit date: | 2013-02-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Selective Class Iia Histone Deacetylase Inhibition Via a Non-Chelating Zinc Binding Group
Nat.Chem.Biol., 9, 2013
|
|
4USF
 
 | | Human SLK with SB-440719 | | Descriptor: | 4-[4-(6-methoxynaphthalen-2-yl)-1H-imidazol-5-yl]pyridine, STE20-LIKE SERINE/THREONINE-PROTEIN KINASE | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
2W52
 
 | | 2 beta-glucans (6-O-glucosyl-laminaritriose) in both donor and acceptor sites of GH16 Laminarinase 16A from Phanerochaete chrysosporium. | | Descriptor: | PUTATIVE LAMINARINASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Vasur, J, Kawai, R, Andersson, E, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2008-12-03 | | Release date: | 2009-07-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-Ray Crystal Structures of Phanerochaete Chrysosporium Laminarinase 16A in Complex with Products from Lichenin and Laminarin Hydrolysis
FEBS J., 276, 2009
|
|
7AMA
 
 | | IL-17A in complex with small molecule modulators | | Descriptor: | Interleukin-17A, ~{N}-[(2~{S})-1,1-dicyclopropyl-3-[[4-(3,5-dimethyl-1~{H}-pyrazol-4-yl)phenyl]amino]-3-oxidanylidene-propan-2-yl]-2-propan-2-yl-pyrazole-3-carboxamide | | Authors: | Hakansson, M, Kimbung, R, Logan, D, Walse, U.B, de Groot, M.J, Andrews, M.D, Dack, K.N, Lambert, M. | | Deposit date: | 2020-10-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of an Oral, Rule of 5 Compliant, Interleukin 17A Protein-Protein Interaction Modulator for the Potential Treatment of Psoriasis and Other Inflammatory Diseases.
J.Med.Chem., 65, 2022
|
|
1K36
 
 | | NMR Structure of human Epiregulin | | Descriptor: | Epiregulin | | Authors: | Sato, K, Miura, K, Tada, M, Aizawa, T, Miyamoto, K, Kawano, K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-09-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Febs Lett., 553, 2003
|
|
7AMG
 
 | | IL-17A in complex with small molecule modulators | | Descriptor: | (3~{R})-4-[4-[[(2~{S})-2-[[2,2-bis(fluoranyl)-2-phenyl-ethanoyl]amino]-3-(2-chlorophenyl)propanoyl]amino]phenyl]-3-[[(2~{S})-3-methyl-2-[2-[2-[(2-methylpropan-2-yl)oxycarbonylamino]ethoxy]ethanoylamino]butanoyl]amino]butanoic acid, Interleukin-17A | | Authors: | Hakansson, M, Kimbung, R, Logan, D, Walse, U.B, de Groot, M.J, Andrews, M.D, Dack, K.N, Lambert, M. | | Deposit date: | 2020-10-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Discovery of an Oral, Rule of 5 Compliant, Interleukin 17A Protein-Protein Interaction Modulator for the Potential Treatment of Psoriasis and Other Inflammatory Diseases.
J.Med.Chem., 65, 2022
|
|
2VTM
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, PYRAZOLO[1,5-A]PYRIMIDINE-3-CARBONITRILE | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VU3
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-20 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
4RGJ
 
 | | Apo crystal structure of CDPK4 from Plasmodium falciparum, PF3D7_0717500 | | Descriptor: | Calcium-dependent protein kinase 4 | | Authors: | Wernimont, A.K, Walker, J.R, Hutchinson, A, Seitova, A, He, H, Loppnau, P, Neculai, M, Amani, M, Lin, Y.H, Ravichandran, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lovato, D.V, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-30 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Apo crystal structure of CDPK4 from Plasmodium falciparum, PF3D7_0717500
To be Published
|
|
1K37
 
 | | NMR Structure of human Epiregulin | | Descriptor: | Epiregulin | | Authors: | Sato, K, Miura, K, Tada, M, Aizawa, T, Miyamoto, K, Kawano, K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-09-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of epiregulin and the effect of its C-terminal domain for receptor binding affinity
Febs Lett., 553, 2003
|
|
2IK8
 
 | | Crystal structure of the heterodimeric complex of human RGS16 and activated Gi alpha 1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Soundararajan, M, Turnbull, A.P, Papagrigoriou, E, Debreczeni, J, Gorrec, F, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-02 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2W39
 
 | | Glc(beta-1-3)Glc disaccharide in -1 and -2 sites of Laminarinase 16A from Phanerochaete chrysosporium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-glucono-1,5-lactone, PUTATIVE LAMINARINASE, ... | | Authors: | Vasur, J, Kawai, R, Andersson, E, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2008-11-07 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-Ray Crystal Structures of Phanerochaete Chrysosporium Laminarinase 16A in Complex with Products from Lichenin and Laminarin Hydrolysis
FEBS J., 276, 2009
|
|
2VPZ
 
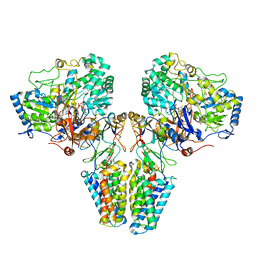 | | Polysulfide reductase native structure | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, HYPOTHETICAL MEMBRANE SPANNING PROTEIN, IRON/SULFUR CLUSTER, ... | | Authors: | Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T, Curmi, P, Iwata, S. | | Deposit date: | 2008-03-09 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism of Energy Conservation in Polysulfide Respiration
Nat.Struct.Mol.Biol., 15, 2008
|
|
7BS8
 
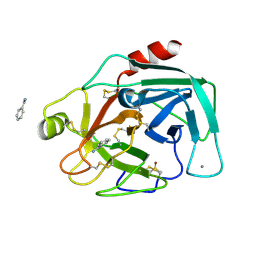 | | Bovine Pancreatic Trypsin with Benzamidine (Cryo) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
2IEZ
 
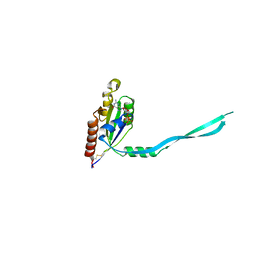 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7BS2
 
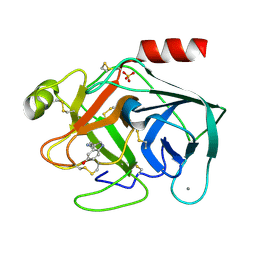 | | Bovine Pancreatic Trypsin with serotonin (Room Temperature) | | Descriptor: | CALCIUM ION, Cationic trypsin, SEROTONIN, ... | | Authors: | Maeki, M, Ito, S, Takeda, R, Funakubo, T, Ueno, G, Ishida, A, Tani, H, Yamamoto, M, Tokeshi, M. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Room-temperature crystallography using a microfluidic protein crystal array device and its application to protein-ligand complex structure analysis.
Chem Sci, 11, 2020
|
|
7TBL
 
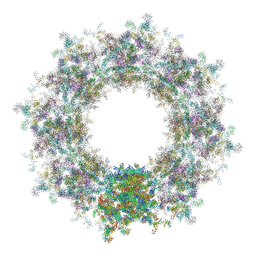 | | Composite structure of the human nuclear pore complex (NPC) cytoplasmic face generated with a 12A cryo-ET map of the purified HeLa cell NPC | | Descriptor: | DDX19, ELYS, GLE1, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
2J6D
 
 | | CONKUNITZIN-S2 - CONE SNAIL NEUROTOXIN - DENOVO STRUCTURE | | Descriptor: | CONKUNITZIN-S2 | | Authors: | Korukottu, J, Bayrhuber, M, Montaville, P, Vijayan, V, Jung, Y.-S, Becker, S, Zweckstetter, M. | | Deposit date: | 2006-09-27 | | Release date: | 2007-01-16 | | Last modified: | 2021-06-23 | | Method: | SOLUTION NMR | | Cite: | Fast High-Resolution Protein Structure Determination by Using Unassigned NMR Data.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2ICP
 
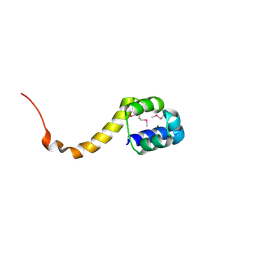 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 4.0. Northeast Structural Genomics Consortium TARGET ER390. | | Descriptor: | MAGNESIUM ION, antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the bacterial antitoxin HigA from Escherichia coli.
To be Published
|
|
