5DVW
 
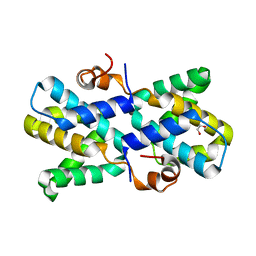 | |
3QHX
 
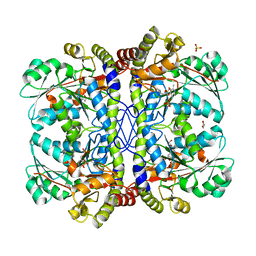 | |
5EQZ
 
 | |
7U0U
 
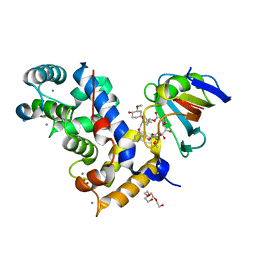 | | Crystal Structure of a Aspergillus fumigatus Calcineurin A - Calcineurin B fusion bound to FKBP12 and FK-506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fox III, D, Abendroth, J, DeBouver, N.D, Hoy, M.J, Heitman, J, Lorimer, D.D, Horanyi, P.S, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Synthesis of FK506 and FK520 Analogs with Increased Selectivity Exhibit In Vivo Therapeutic Efficacy against Cryptococcus.
Mbio, 13, 2022
|
|
5DLE
 
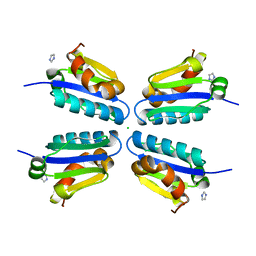 | |
5ENU
 
 | |
5ER6
 
 | |
5W8I
 
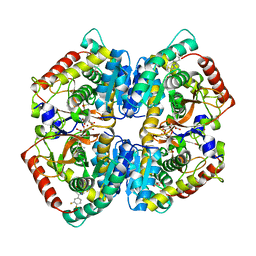 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 23 and Zinc | | Descriptor: | 2-[3-(3,4-difluorophenyl)-5-hydroxy-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Abendroth, J. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
3SX2
 
 | |
6ONA
 
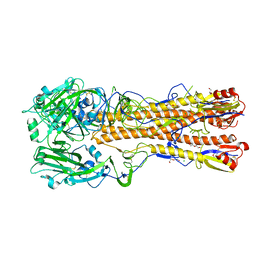 | |
4J5I
 
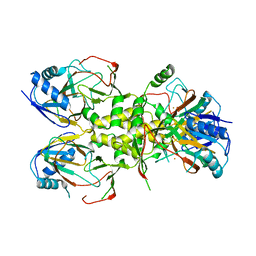 | |
3V9O
 
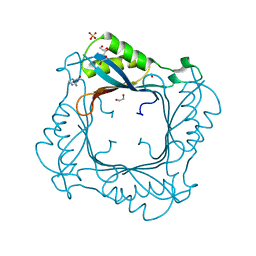 | |
4PFS
 
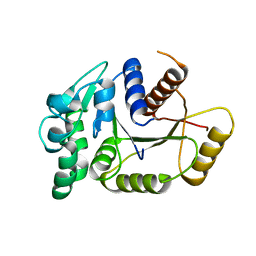 | |
3V9P
 
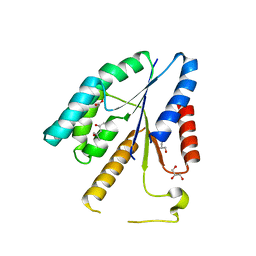 | |
4LGV
 
 | |
4P9A
 
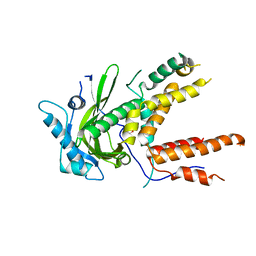 | | X-ray Crystal Structure of PA protein from Influenza strain H7N9 | | Descriptor: | GLYCEROL, Polymerase PA | | Authors: | Fairman, J.W, Moen, S.O, Clifton, M.C, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Structural analysis of H1N1 and H7N9 influenza A virus PA in the absence of PB1.
Sci Rep, 4, 2014
|
|
3UPT
 
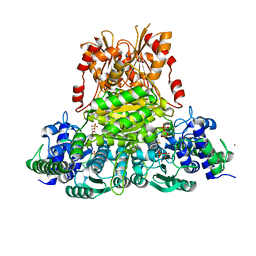 | |
6VZZ
 
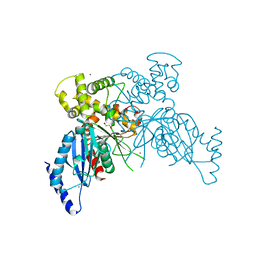 | |
4KAM
 
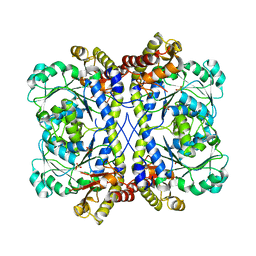 | |
3HZG
 
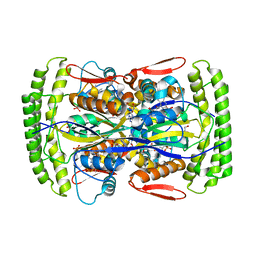 | | Crystal structure of mycobacterium tuberculosis thymidylate synthase X bound with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Staker, B.L, Rathod, P, Hunter, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increasing the structural coverage of tuberculosis drug targets.
Tuberculosis (Edinb), 95, 2015
|
|
3NWO
 
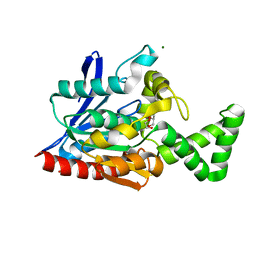 | |
3OC7
 
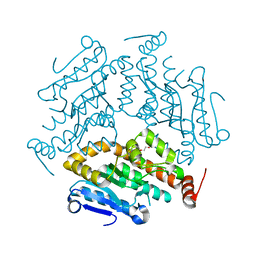 | |
3MOY
 
 | |
3NDN
 
 | |
5C9C
 
 | | CRYSTAL STRUCTURE OF BRAF(V600E) IN COMPLEX WITH LY3009120 COMPND | | Descriptor: | 1-(3,3-dimethylbutyl)-3-{2-fluoro-4-methyl-5-[7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl]phenyl}urea, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Edwards, T, Abendroth, J, Chun, L. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers.
Cancer Cell, 28, 2015
|
|
